1. Introduction
Molecular studies significantly depend on the extraction of high-quality DNA to ensure its suitability for subsequent downstream applications, such as DNA genotyping1. Single nucleotide polymorphisms (SNPs) are sequence variations caused by a single nucleotide mutation at a specific locus in the DNA2. Currently, SNP markers are one of the preferred genotyping approaches due to their abundance in the genome, genetic stability, and compatibility with high-throughput automated analysis3. SNP genotyping technologies serve as powerful resources for investigating quantitative genetics, population genetics and molecular evolution4. Therefore, ensuring a robust DNA extraction method is essential for obtaining genomic DNA with both high quality and yield, crucial for accurate SNP genotyping.
Different biological samples, such as blood, ear tissue and semen, are suitable for sheep DNA analysis using conventional manual techniques. However, these materials vary in terms of ease of collection, storage requirements, and necessary shipment conditions. Furthermore, while numerous studies comparing DNA extraction from various sources have been published in cattle 5)(6)7)(8) , fewer studies focus on sheep 9)(10) 11.
Although blood remains the primary source of genomic DNA in sheep due to its reliability in providing high-quality DNA, blood collection may induce stress in animals, and proficient technicians are essential to ensure proper sample collection. Another drawback is the need to preserve blood at 4°C for shipment and storage. Semen provides an alternative method for DNA collection, but its applicability is primarily limited to high-genetic-value rams. Ear punching, in addition to being an invasive procedure, requires preservation, typically achieved using alcohol. In contrast, nasal swabbing, a technique predominantly used in humans12, offers the advantages of non-invasiveness, ease of sampling, and room temperature storage, making it an appealing option alongside traditional collection methods.
Identifying a non-invasive and straightforward technique for obtaining high quality DNA is critical to ensure successful sampling at the farm level. Thus, in this study, we assessed and compared two biological matrices -nasal swabs and blood- for the isolation of genomic DNA from sheep. Our goal was to determine the suitability of nasal swabs as a biological matrix to obtain high-quality DNA suitable for SNP genotyping in sheep.
2. Materials and methods
2.1 Sample collection
Blood samples were collected from 15 female Texel lambs raised at INIA´s (National Agricultural Research Institute) Animal Production Unit, an experimental farm located at INIA Las Brujas (Canelones, Uruguay). Peripheral blood samples were obtained through jugular vein puncture using 6 ml K2 EDTA blood collection tubes (BD, New Jersey, US). These samples were gently inverted to prevent clotting and immediately placed in insulated boxes with ice. Once in the laboratory, buffy coat was extracted from blood by spinning whole blood at 3500 RPM (1300 xg) for 10 min in an Eppendorf (5415R) centrifuge at 4°C. Buffy coat was removed, mixed with 1 ml of red blood cell lysis buffer (0.14 M NH4Cl, 17 mM Tris base) and centrifuged at 8000 RPM (6797 xg) for 1 min at 4°C. The liquid was discarded, and the process was repeated until a clean buffy coat was obtained.
Nasal swabs were collected from the same 15 Texel lambs using standard commercial nasal swabs (Johnson & Johnson, New Jersey, US). The nasal swab sponge was gently rubbed inside each animal´s nostril for 3 s, ensuring it did not extend beyond a depth of 1 cm. Subsequently, the swabs were air-dried for 5 min. Following this, the swabs were carefully placed in envelopes and stored at room temperature.
2.2 DNA extraction
We employed a single DNA extraction method for both matrices13: buffy coat from blood and nasal swab. For the buffy coat, leucocytes were suspended in 600 µl of digestion buffer (10 mM Tris-HCl (pH 8.0), 0.4 M NaCl, 2% SDS, 50 mM EDTA), and 5 µl of proteinase K (20 mg/ml, Meridian Bioscience, Cincinnati, OH) were added. The mixture was incubated overnight at 55°C. The following day, 5 µl of RNase A (10 mg/ml, Fermentas, Waltham, MA) were added to the lysate and incubated at 37°C for 30 min. Subsequently, 300 µl of potassium acetate 3 M were added and left at -20°C for 10 min. After centrifugation at 13000 RPM (17949 xg) for 10 min, 600 µl of the supernatant were transferred to a 1.5 ml tube, and 500 µl of isopropanol were added. After 20 min at -20°C, the mixture was centrifuged at 13000 RPM (17949 xg) for 10 min and the supernatant was discarded. The pellet was washed with 600 µl of 70% ethanol, and after centrifugation at 13000 RPM (17949 xg) for 5 min, the supernatant was discarded. The pellet was washed again with 100% ethanol. Following a final centrifugation, the DNA pellet was dried and re-suspended in 100 µl elution buffer (10 mM Tris-HCl, pH=8.0).
For the nasal swab, the sponge was cut inside a 1.5 ml tube, and the extraction procedure employed for the buffy coat was replicated, with the following adjustments in the quantities of certain reagents: 700 µl of digestion buffer, 200 µl of potassium acetate 3 M, and 50 µl of elution buffer.
2.3 Spectrophotometer measurements
Conducting spectrophotometric measurements of ultraviolet light absorbance at various wavelengths (260 nm and 280 nm) provides a quick and efficient method for the initial assessment of purity and concentration in nucleic acid samples14. In this study, both quantity and quality were assessed using a NanoDrop 8000 spectrophotometer (Thermo Scientific, Waltham, MA). DNA concentration was determined by utilizing the absorbance reading at 260 nm through the application of the Beer-Lambert law15. Additionally, the 260/280 nm absorbance ratio (A260/A280 ratio) was computed to assess protein contamination, with values falling within the 1.8-2.0 range considered optimal14. Total DNA yield was calculated by multiplying extracted DNA concentration by elution volume.
2.4 Gel electrophoresis
DNA integrity was evaluated through gel electrophoresis, where 1 µl of each DNA extract was analyzed in a 1% agarose gel containing 0.5X TBE buffer (Tris-Borate-EDTA). The electrophoresis was conducted for 25 min at 100V, and the results were visualized under UV illumination.
2.5 SNP genotyping
SNP genotyping was performed using the Applied Biosystems™ Axiom™ Ovine Genotyping Array 50K (Thermo Fisher Scientific, Waltham, MA, US) at Genexa Laboratory (Montevideo, Uruguay). Genomic data quality control was conducted using QCF90 software16, which involved the removal of SNPs located in sexual chromosomes, monomorphic and those with minor allele frequencies (MAF) <0.05, with call rate <90%, and significant deviation from HWE (Hardy-Weinberg Equilibrium) test at p<10-6. Additionally, animals with call rate <90% were removed. Following quality control, 30,302 SNPs remained for further analysis.
2.6 Animal Welfare
Animal handling and blood collection procedures were approved by the Animal Ethics Committee of INIA (approval number INIA_2018.2; unreferenced), in accordance with Uruguayan laws for the care and management of experimental animals (Law 18611)17.
2.7 Statistical Analysis
DNA concentration, DNA yield, and A260/A280 ratio obtained from both blood and nasal swabs were evaluated for normal distribution using the Shapiro-Wilk test, while equality of variances was assessed with the Levene's test. DNA concentration and DNA yield showed a normal distribution and homogeneity of the variances; therefore, a paired Student t test implemented in R12 was applied to compare the means for these variables. The A260/A280 ratio also showed a normal distribution, but the variances were not equal between the groups; therefore, we applied a Wilcoxon test to assess significant differences, also using R18.
Genotype quality was assessed by the call rate per animal. Call rate is defined as the ratio of genotypes passing the calling threshold to the total number of genotypes that require inference19 with values ranging from 0 to 100%. Higher values are preferred.
Concordance rates between genotypes obtained from blood and nasal swab matrices were assessed using SeekParentF90 software20. Out of the initial 15 samples, four nasal swab samples failed genotyping, leading to their exclusion from the concordance analysis. Despite exhibiting concentrations in the lower range (69.4 to 110.7 ng/µl and a total yield between 3.5 and 5.5 µg), these concentrations surpassed the minimum requirement for SNP genotyping at Genexa Laboratory.
3. Results
3.1 DNA concentration, spectrophotometer measurements and total DNA yield
The two biological matrices yielded genomic DNA of varying quality and quantity (Table 1). For DNA concentration, the difference between mean values for blood (159.14 ng/µl) and nasal swab (130.12 ng/µl) matrices was non-significant (p>0.05).
On the other hand, significant differences were detected for A260/A280 ratio and total DNA yield (p<0.05), with blood matrix showing a slightly lower ratio (1.90) and a higher yield (15.91 µg). Mean A260/A280 ratios for both samples were within the 1.8-2.0 range, which indicates the presence of pure DNA9.
Table 1: Comparison of mean DNA concentration (Conc), A260/A280 ratio and total DNA yield obtained from two different biological matrices, with standard errors in parentheses
| Matrix | Conc. (ng/µl) | A260/A280 ratio | Total yield (µg) |
|---|---|---|---|
| Blood | 159.14(10.89)a | 1.90(0.00)a | 15.91(1.09)a |
| Nasal swab | 130.12(15.32)a | 1.96(0.01)b | 6.51(0.77)b |
a,b Comparison of values within each column; values with the same superscript are not statistically different (p>0.05) from each other but they differ significantly (p<0.05) from values with different superscript.
3.2 Gel electrophoresis
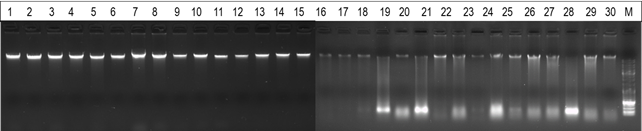
Integrity of extracted genomic DNA was assessed by agarose gel electrophoresis (Figure 1). Gel electrophoresis analysis revealed that DNA extracted from blood samples exhibited a higher degree of integrity, evident through well-defined and concentrated bands. In contrast, DNA obtained from nasal swabs displayed some degree of smearing, indicative of DNA fragmentation.
3.3 Call rates and concordance analysis
In terms of call rates, both blood and nasal swabs yielded similar results. The mean call rate per individual obtained using blood (0.995) was slightly higher than the one obtained using nasal swab (0.994), but the difference was non-significant (p>0.05).
Genotyping data was compared to assess concordance rate between blood and nasal swab matrices (Table 2). High concordance rates were found, ranging from 0.898 to 0.999, with a mean of 0.984 and a standard error of 0.10. These results provide evidence that genotyping results using DNA isolated from both blood and nasal swabs show a substantial level of similarity.
Table 2: Genotype concordance rates (ratio) comparing blood matrix (Id_1) and nasal swab matrix (Id_2), call rates obtained with blood matrix (callrate_1), and call rates obtained with nasal swab matrix (callrate_2)
| Animal Id | Id_1 | Id_2 | ratio | callrate_1 | callrate_2 |
|---|---|---|---|---|---|
| 139020200029 | 20210409069 | 20220810071 | 0.998 | 0.999 | 0.998 |
| 139020200171 | 20210409075 | 20220810072 | 0.995 | 0.995 | 0.999 |
| 139020200215 | 20210409076 | 20220810073 | 0.997 | 0.998 | 0.999 |
| 139020200276 | 20210409078 | 20220810074 | 0.948 | 0.999 | 0.951 |
| 139020200277 | 20210409079 | 20220810075 | 0.998 | 0.999 | 0.999 |
| 139020200294 | 20210824013 | 20220810076 | 0.997 | 0.998 | 0.999 |
| 139020200324 | 20210409082 | 20220810077 | 0.898 | 0.964 | 0.999 |
| 139020200363 | 20210409086 | 20220810078 | 0.997 | 0.999 | 0.997 |
| 139020200369 | 20210824015 | 20220810079 | 0.999 | 0.999 | 0.999 |
| 139020200391 | 20210824018 | 20220810080 | 0.997 | 0.996 | 0.999 |
| 139020200401 | 20210824023 | 20220810081 | 0.998 | 0.998 | 0.999 |
4. Discussion
DNA concentration obtained from blood samples in our study exceeded the average amount reported by Nuñez and others11, who used 50 µl of blood and a proteinase K protocol, yielding a concentration of 42.45 ng/µl. Our findings closely resembled the yield reported by Psifidi and others10, who used 10 ml of blood and a commercial kit (162 ng/µl), and Foley and others7, who used 450 µl of blood with a proteinase K protocol (158.2 ng/µl). However, in the case of nasal swabs, a previous study7 reported higher DNA concentrations (150.9 ng/µl) compared to our study. It's noteworthy that these authors used commercial nasal swabs (Performagene) containing a solution that acts as a preservative and cell lysis agent, whereas we employed regular nasal swabs. Consequently, our study reported lower total DNA yield values compared to Foley and others7 for nasal swabs (15.09 µg).
Concerning mean A260/A280 ratios, our results exhibited superior ratios compared to those reported by Neary and others8 for both blood (1.6) and nasal swabs (1.75), as well as by Foley and others7 for blood (1.7).
In comparison to a previous study7, our study showed a similar high degree of DNA integrity, characterized by well-defined bands in blood samples. However, this was not observed in nasal swab samples. Once again, this discrepancy could be attributed to the preservation solution used in the commercial swabs, which may have caused DNA fragmentation.
Even though DNA extracted from blood matrix exhibited superior performance, DNA derived from nasal swabs demonstrated satisfactory results, yielding high quantity (130.12 ng/µl), optimal A260/A280 ratio (1.96), and sufficient total yield (6.51 µg) suitable for SNP genotyping. Moreover, nasal swabs provide additional benefits, including ease of use (requiring minimal expertise and equipment for sample collection), accessibility (commercial swabs commonly found in stores), and the convenience of storing samples at room temperature. This confers a notable advantage over the use of blood samples, particularly for sampling in commercial farms. However, given the relatively high failure rate of 27% in nasal swab samples during genotyping, there is a need to revise and enhance the DNA extraction protocol from nasal swabs.
5. Conclusions
In conclusion, we observed a high concordance rate between genotypes obtained from blood and nasal swabs in this study. These results suggest that nasal swabs are a viable alternative to obtain high quality sheep DNA suitable for SNP genotyping at large scale. However, further studies are needed to improve the DNA extraction protocol from nasal swabs.