Introduction
Sorghum cultivation (Sorghum bicolor (L.) Moench) in Uruguay covers a total planting area of 47,000 hectares and a production of 214,000 tons1. This figure only considers dry grain sorghum harvest, wet grain or forage sorghum are not taken into account, so it is estimated that the crop would cover thousands of hectares more in the country.
In January 2017, a sample of sorghum, cultivar F1497, with concentric necrotic lesions on the leaves enters the laboratory. Symptomatology is associated with the sorghum disease called zonate leaf spot caused by the fungus Microdochium sorghi (d.c. Bain & Edgerton ex Deighton) U. Braum (sin. Gloeocercospora sorghi d.c. Bain & Edgerton ex Deighton, Ramulispora andropogonis Miura, Titaeospora andropogonis (Miura) f.l. Tai). In the same growing season, the disease was observed in 29%of the evaluated materials in the sorghum official trials of Evaluación Nacional de Cultivares inase-inia at La Estanzuela and Young2.
Zonated leaf spot is common in wet areas of southern USA3 and may affect other crops, such as corn, sugar cane and numerous pastures3)(4. The pathogen is present in South America: Argentina, Brazil, and Venezuela5, but it has not been reported in Uruguay. The objective of this study was to identify and report the causative agent of this leaf spot in our country.
Material and Methods
Isolation
The pathogen was isolated from conidia produced in the sporodochia on the upper side of the leaf with a loop, and transferred to Petri dishes containing PDA (extract of 200 g potato, 10 g dextrose, 20 g agar in 1000 ml of dH2O).
Identification
The identification was carried out by observing the characteristics of the lesion and the sporodochia, and measuring conidia under a microscope. In addition, growth and morphology of the structures in PDA culture medium were observed.
Genomic dna was extracted for molecular identification with DNeasy® Plant Mini Kit (Qiagen Inc.). The amplification was performed by conventional pcr using the primers ITS 5 (52 -ggaagtaaaagtcgtaacaagg) and ITS 4 (52 -tcctccgcttattgatatgc)6. The mixing and cycling conditions were carried out according to Abreo and others7. pcr products were sequenced using the standard service of Macrogen Inc., South Korea (http://www.macrogen.com). The obtained sequence was manually corrected using the program MEGA version 68 and aligned with similar sequences obtained from GenBank9. A phylogenetic analysis was performed by the maximum parsimony method with the MEGA software10. The new isolate was deposited into INIA Las Brujas’ collection with the code ilb 356.
Pathogenicity Tests
The fungus was inoculated in Petri dishes with PDA. Ten days later, the medium containing microsclerotia and mycelium was macerated, passing it several times through a syringe. The paste that resulted from the maceration of three plates was diluted in 100 ml of sterile water. Four pots with 5 plants, each of forage sorghum cultivar Estanzuela Comiray, were inoculated by spraying at third leaf sheath visible stage, covered with nylon bags for 24 hours and kept in a growth chamber at 24 °C. Two sorghum pots were used as control, with the same treatment, except that the spraying was performed with macerated PDA without the fungus. Once the lesions developed, the fungus was reisolated from the inoculated plants.
Results
Symptomatology and identification
Leaf lesions were circular or semicircular, alternating purple to red bands with brown to cinnamon bands, resulting in a concentric or zonate lesion (Figure 1). Several coalescing lesions affected most of the leaf surface.
The fungus produces a lot of conidia over the lesions, which emerge from stomata as a pink gelatinous mass, product of sporodochia sporulation. Under a microscope, long filiform, curved conidia of variable size between 1.5-3.5 x 20-190 µm were observed, presenting up to 10 septa. On senescent tissue and PDA medium, black microsclerotia with 0.1 to 0.2 mm in diameter were observed.
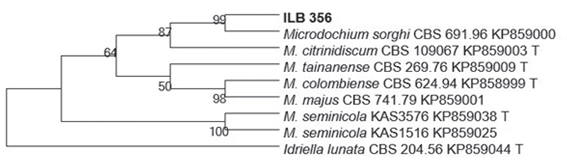
Phylogenetic analysis, based on the alignment of GenBank related sequences, allowed to group ilb 356 strain with an isolate of Microdochium sorghi (accession KP85900)11 with a Bootstrap value of 99 %, sharing 521 of the 522 bp (Figure 2). The ITS sequence was deposited in GenBank with the number MH38491012.
Pathogenicity Tests
Three days after inoculation small aqueous lesions were observed on the leaves. Five days later, typical lesions of the disease were observed on the leaves (Figure 3), while the control did not develop symptoms.
The pathogen reisolated from the inoculated leaves presented the same morphological characteristics as the inoculated isolate.
Conclusion
The observed symptomatology coincides with the description of the sorghum disease called zonate leaf spot4)(13. The symptoms were reproduced in plants inoculated in growth chamber and the fungus was reisolated from the lesions, fulfilling Koch postulate. The morphological characters and the molecular analysis allowed to confirm the identity of the pathogen causing the zonate spot as Microdochium sorghi. It should be noted that, although the taxonomy and phylogeny of this fungal group were revised11, this sorghum pathogen is still named in literature as Gloeocercospora sorghi13. Although symptoms had been previously observed in the field2, this study represents the first laboratory-confirmed report of the causal pathogen of zonate leaf spot in Uruguay.