1. Introduction
Ralstonia solanacearum is one of the world’s most important phytopathogenic bacteria due to its lethality, persistence in the fields, wide host range, and broad geographical distribution1)(2. This soil-borne vascular pathogen causes bacterial wilt (bw) in more than 250 monocot and dicot plant species in tropical, subtropical and temperate regions3. R. solanacearum is considered a species complex composed by a heterogeneous group of related strains classified in four lineages termed phylotypes based on their phylogeography4.
A recent taxonomic revision has led to the distinction of three separate species within the R. solanacearum species complex. Based on this new classification, the species R. solanacearum includes strains from phylotype II, originated from the southern Americas. Phylotypes I and III were assigned to the taxonomic species R. pseudosolanacearum and phylotype IV has been reclassified as R. syzygii5.
R. solanacearum infects the roots of host plants, rapidly colonizes the vascular system and releases large amounts of exopolysaccharides which obstruct the water flow within xylem vessels, causing wilting symptoms and subsequent plant death6. The pathogen can alternate hosts and persist, spread, and survive in different natural habitats, including soil, weed, plant debris, rhizospheres. In addition, it can be spread through irrigation water or infected planting material hindering disease control7)(8)(9. This set of characteristics makes it very challenging to achieve a sustainable management of the disease10.
Potato (Solanum tuberosum L.) is the third most important food crop after rice and wheat and is consumed by more than a billion people worldwide11. bw caused by R. solanacearum has been estimated to affect about 1.7 million hectares of potatoes in approximately 80 countries, with usd 950 million of global damage yearly12. Phylotype iib sequevar 1 (iib-1), formerly known as race 3/biovar 2A, causes bw of potato in over 90% of this crop in cold and temperate regions or high altitudes in the tropics13)(14)(15.
Improving plant resistance is the most sustainable, cost-effective and environmentally prospect strategy to manage plant diseases. Chemical control is generally ineffective, phytosanitation and cultural measures are difficult to apply, and biological control agents are not commercially available12)(16)(17. For most bw susceptible crops, the sources of high level, gene-for-gene type of resistance encoded by single dominant genes are scarce18. Instead, available sources of resistance are usually polygenic, being difficult to transfer into desirable cultivars. Although quantitative trait loci (qtl) associated with bw resistance have been found in tomato19)(20)(21, tobacco22, eggplant23 and potato24, resistant commercial cultivars are not yet available.
Resistance genes in potato gene pool have been identified in several potato-related wild species. Solanum commersonii Dun has been used as the main wild genetic resource for the potato breeding program at the National Institute of Agricultural Research (inia), Uruguay. This wild relative is characterized by high genetic diversity25)(26 and desirable traits, such as low temperature tolerance and resistance to several pathogens, including R. solanacearum8)(26)(27)(28)(29)(30)(31. Promising accessions and interspecific hybrids with partial resistance to bw were obtained from this program28)(29)(31)(32.
Although resistance has been identified in some wild potato species, introgression of these genes through classical breeding reached only partial resistance and, usually, a poor agronomic performance is also dragged18)(33. A specific way to improve plant disease resistance is to enhance the capability of the plants’ innate immune system34. Besides constitutive physical and chemical barriers, plants also have an active recognition of microbial invaders.
Plants can detect highly conserved microbial molecules, named pathogen- or microbe-associated molecular patterns (pamp or mamp), by pattern recognition receptors (prrs) triggering a series of defense responses. This first defense line named as pamp triggered immunity (pti) includes reactive oxygen species (ros) burst, callose deposition, activation of pathogenesis-related (pr) gene expression, and induction of mitogen-activated protein kinase (mapk) cascades34)(35)(36)(37.
The elongation factor-Tu (ef-tu) receptor (efr) from Arabidopsis thaliana (atefr) is a prr that recognizes the bacterial pamp ef-tu or its eliciting epitope elf1838)(39. The interfamily transfer of atefr has shown to confer elf18 perception and to increase pathogens resistance in several plants. This behaviour was observed in Solanaceous crops such as Nicotiana benthamiana against Agrobacterium tumefaciens, Pseudomonas syringae pv. syringae and P. syringae pv. tabaci, and in tomato against R. solanacearum17)(34. The efr contribution on resistance against bacterial diseases was verified also in monocot species such as rice, against Xanthomonas oryzae pv. oryzae, and wheat, against Pseudomonas syringae pv. oryzae37)(40)(41.
Recently, our group evaluated the effect of atefr gene expression on a commercial potato line (inia Iporá) and an interspecific breeding line (clone 09509.6), carrying quantitative bw resistance introgressed from S. commersonii Dun33. We found that both atefr lines showed greater resistance to R. solanacearum after plant inoculation, under controlled conditions in a growth chamber. Interestingly, complementation of heterologous expression of atefr, with quantitative resistance acquired from the wild potato species, was clearly evidenced in bw resistance. It was concluded that this strategy could be promising to develop resistance against R. solanacearum in potato crop33.
In this work, we further explored the application potential of these potato atefr transgenic lines by evaluating their bw resistance using conditions resembling natural field infection. In addition, the effects of atefr on the R. solanacearum colonization, dissemination and multiplication patterns were evaluated.
We verified that in both potato genetic backgrounds, atefr enhanced the bw resistance under conditions similar to those of field crops, with lower disease symptoms both at the foliar and tuber levels. On the other hand, by studying the bacterial colonization pattern, we discovered that atefr results in restriction and limitation of bacteria at basal stem, which delays or prevents the advance of R. solanacearum to the upper parts of the plant. Moreover, this effect seems more pronounced in the interspecific breeding line, possibly leading to a more effective activation of the plant immune system.
2. Materials and methods
2.1 Bacterial strains and growth conditions
Ralstonia solanacearum strain UY031 (race 3, biovar 2A/phylotype iib, sequevar 1)42 and R. solanacearum reporter strains UY031 Pps-lux and UY031 Pps-gfp43 were grown at 28 °C in Kelman medium supplemented with 2,3,5-trifenil tetrazolium chloride (ttc) for 48-72 h. Gentamicin was used for selection of reporter strains (5 and 75 µg·ml−1 in liquid and solid cultures, respectively).
2.2 Plant material and growth conditions
Solanum tuberosum cv. inia Iporá (susceptible to bw) and the partially resistant breeding clone 09509.6 were used as wild-type controls in this study. Clone 09509.6 is a second backcross (BC2) genotype which harbours partial bw resistance introgressed from S. commersonii Dun29)(44. In addition, four atefr transformation events (Iporá atefr 3, Iporá atefr 12, clone 09509.6 atefr 34 and clone 09509.6 atefr 37) were selected from a previous study33.
Plants were micro-propagated in vitro from nodal stems in Murashige and Skoog (ms) medium with sucrose 30 g.l-1 and kept at 22°C with cycles of 16:8 h light:darkness. After two weeks, plantlets were transferred into multicell trays containing treff soil mix (Treff Substrates bv, Moerdijk, The Netherlands), and grown for two to five weeks in a greenhouse under natural light.
2.3 High tunnel disease assay
For disease evaluation in conditions resembling natural field infection, a high tunnel plastic house which fulfilled biosafety level II regulations for gmo plants was built. It included anti-insect mesh, and three replicates (blocks) filled with an enriched soil substrate, contained in elevated cement seed beds and a disinfection area to prevent outside bacterial dissemination (Figure S1, Supplementary Material). Temperature and relative humidity conditions were recorded (Table S1, Supplementary Material). The average temperature varied between 16.93 and 34.51 °C, and average hr between 45.88 and 87.66%.
Plants were micro-propagated in vitro as described above and grown for five weeks in a greenhouse prior to transplanting in the high tunnel. In these trails, Iporá wild-type, Iporá atefr 3, Iporá atefr 12, clone 09509.6 wild-type, clone 09509.6 atefr 34 and clone 09509.6 atefr 37 were assayed.
Experimental plots consisted of 10 plants per genotype per plot with a 30-cm distance between plants. A randomized complete block design was used with 3 replicate plots per treatment. Two field trials were conducted during the summer season of 2019 and autumn of 2020.
Soil inoculation was adapted from the protocol described by Bonierbale45. Bacterial suspensions were prepared from overnight liquid cultures of R. solanacearum UY031 incubated at 28 °C, and spectrophotometrically adjusted to a concentration of 108 cfu·ml−1 (OD600 of 0.1). Petri dishes (9 cm) containing ttc Kelman medium were inoculated with 100 µL of this suspension employing the spread plate technique, with the aim of achieving confluent growth throughout the plate. After 48 h incubation, a piece (1/8) of grown agar plate was buried at 5 cm distance from each plant and 20 cm deep45.
Disease progression was registered regularly using an ordinal scale ranging from 0 (asymptomatic plant) to 4 (all leaves wilted). Weekly evaluations were performed until all plants from the susceptible cultivar inia Iporá died, at 60 days after inoculation (dai). Tubers from all plants were harvested and visually inspected for external or internal symptoms of bacterial wilt infection (white secretion exuding from eyes or vascular ring of tubers, especially in the area around the stolon).
Resistance level was calculated by the area under disease progress curve (audpc) based on the average wilt scoring for each plant. Analysis of variance (anova) was performed to identify significant effects of the atefr transgene, the genetic background, and the interaction between the main effects on bw resistance. For each data set, the assumptions of normality and homogeneity of variances were previously verified. Means were compared using Tukey’s multiple comparison test at the 95% confidence level. Statistical analyses were performed using R 3.6.146.
2.4 Plant inoculation with R. solanacearum reporter strains
For colonization pattern evaluation, plants grown in a greenhouse for two weeks were transplanted into individual pots and placed in a growth chamber at 24 °C, with 60% daily/100% night relative humidity, and a photoperiod of 16 h light/8 h darkness for one additional week prior to inoculation assays.
Bacterial suspensions were prepared from overnight liquid cultures of R. solanacearum reporter strains and spectrophotometrically adjusted to a concentration of 107 cfu·ml−1 as described above. Plants were soil inoculated by drenching 40 ml of the bacterial suspension into each pot to reach a final density of 106 cfu·g-1 with previous root wound, as described by Ferreira and others32. Control plants only inoculated with saline solution were considered as negative control treatment.
2.5 Luminescent bacteria visualization
For luminescence detection, 12 plants from each genotype: Iporá wild-type, Iporá atefr 3, Iporá atefr 12, clone 09509.6 wild-type, clone 09509.6 atefr 34, and clone 09509.6 atefr 37 were soil inoculated with the reporter strain UY031 pps-lux. Plants were analyzed using an Invivo msfx pro Bruker system, taking luminescence images with a 5-minute exposure time and X-ray images, evaluating bacterial colonization at 3, 4, 5, 6 and 10 dai.
2.6 Fluorescent bacteria visualization
For fluorescence detection, Iporá wild-type, Iporá atefr 3, Iporá atefr 12, clone 09509.6 wild-type, clone 09509.6 atefr 34, and clone 09509.6 atefr 37 plants were soil inoculated with the reporter strain UY031 pps-gfp. Then, bacterial colonization was analyzed in root and stems tissues at 2, 7 and 14 dai. Roots and transversal sections of lower stem were prepared and fixed as described by Ferreira and others32 and observed in a confocal microscope (Leica, tcs-sp5). Three replicates for each time and genotype were assessed. Experiments were repeated twice.
3. Results and discussion
3.1 Modified atefr potato plants showed enhanced bw resistance in conditions resembling natural field infections
Plants grown in pots under growth chamber conditions do not necessarily behave the same as crops grown in the field. This is because of longer growing cycle and infection times, larger plant size, and the occurrence of several uncontrolled variables, such as temperature, light, humidity, water, pests and nutrients47. Therefore, we decided to study the atefr effect against bw on potato plants grown under fully developmental conditions.
Disease evaluation in the field is challenging due to uneven pathogen concentration in naturally infected soils. Moreover, since R. solanacearum is able to persist in the soil for a long time, artificial field inoculation is no longer possible due to the high dissemination risk. To overcome these limitations, a high tunnel was built allowing uniform experimental conditions and mimic natural field environment while complying with biosecurity measures. Additionally, use of high tunnel minimizes the risk of frost and pests, and allows to reach higher temperatures, promoting disease resistance screening.
A significant effect of the atefr gene (p<0.0001) and the genetic background (p=0.0002) on bw resistance was verified under these experimental conditions, while no significant interaction was observed between both main factors (p=0.2511). According to this result, the two sources of resistance act in an additive way, allowing these main effects to be analyzed independently. The direct effect of atefr gene on BW resistance was verified in the two genetic backgrounds evaluated. Based on audpc calculation, Iporá and 09509.6 atefr transgenic lines were significantly more resistant than their analogue wild-types (Figure 1A, 1B). Regarding Iporá plants, Iporá atefr 12 was the most resistant genotype, displaying higher resistance even when compared to Iporá atefr 3 (p<0.0001). No differences were observed between transgenic lines 09509.6 atefr 34 and 37, showing both significant higher bw resistance compared to the non-transformed clone 09509.6 (p<0.0001).
Differences in bw resistance are also reflected in the symptoms observed for each genotype at the end of the trial (Figures 1C-1H). As expected, plants of the susceptible cultivar inia Iporá wild-type were the first to exhibit wilting symptoms, and all replicate plants were dead from 60 days after inoculation (Figure 1C).
The transgenic lines Iporá atefr 3 and 12 showed 33% and 50% of plant survival, respectively (Figures 1D, 1E). The most relevant effect was observed for plants derived from clone 09509.6 (Figures 1F-1H). In this case, reduced disease symptoms and four-time higher survival rates were observed for both 09509.6 atefr lines compared to the non-transformed clone 09509.6.
After completing the symptoms follow-up, tubers from high tunnel assays were harvested and analyzed for bw symptoms (Figure 2A). At this stage, a high correlation between foliage and tuber symptoms occurrence was observed. Interestingly, atefr plants showed a lower proportion of symptomatic tubers compared to the wild-type controls, although this difference was significant only for the 09509.6 genotypes (Figure 2B).
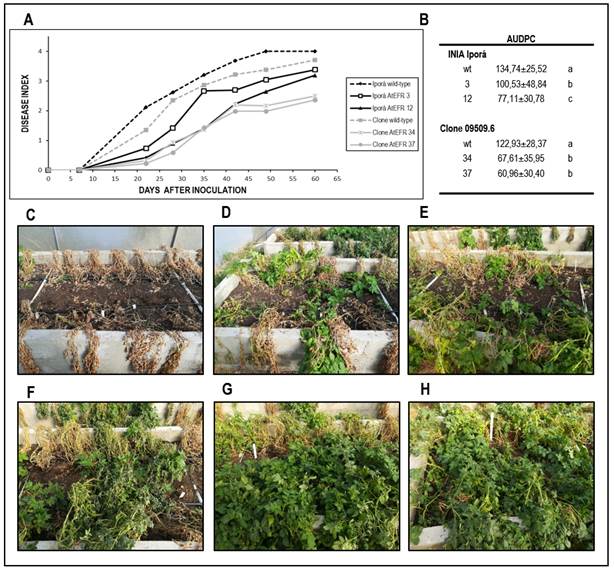
Figure 1: Bacterial wilt (bw) resistance evaluation by high tunnel assays using conditions similar to natural field infection. (A) bw progress curves on Iporá, clone 09509.6 and their atefr transformation events after soil inoculation with Ralstonia solanacearum strain UY031. (B) Area under the disease progress curve (audpc) values for the average wilting scores ± sd as means of two independent experiments. Values followed by the different letters are significantly different (Tukey’s hdl multiple comparison test, p < 0.05). (C-H) Representative experimental plots of Iporá wild-type (C), Iporá atefr 3 (D), Iporá atefr 12 (E), clone 09509.6 wild-type (F), clone 09509.6 atefr 34 (G), and clone 09509.6 atefr 37 (H) at 60 days after inoculation (dai).
These results reflect that the response of atefr potato plants toward bw persists up to tuberization. Even though detection of latent infection was not carried out in this work, we have previously verified that these atefr genotypes exhibit a lower proportion of latently infected tubers33.
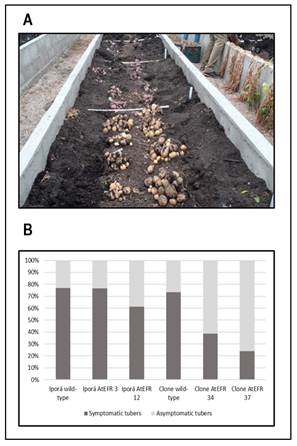
Figure 2: Harvesting of tubers from high tunnel assays using conditions similar to natural Ralstonia solanacearum field infection. (A) Tubers were harvested 74 dai from all experimental plots, and incidence of bw symptoms in tubers was determined for wild-type controls and atefr-transformed lines. (B) Bars represent the number of tubers harvested from 12 randomly chosen plants from each genotype. Tubers with bw symptoms are represented in dark gray, while asymptomatic tubers are denoted in light gray. Similar results were obtained from two repeated independent experiments but only results obtained from the last trial (autumn, 2020) are shown.
In this study, we demonstrate that atefr potato plants show better behavior when bw is present in conditions resembling those in the field. This approach could be a promising alternative to minimize potato crop bacterial wilt infections. Recently, atefr expressed in tomato was shown to be effective in the field as well17. Genetic engineering of pamp recognition has several advantages over the current alternatives to improve resistance to phytopathogens. Using the plant’s own immune system to combat plant diseases should reduce agrochemical inputs and their associated financial, health and environmental costs34. In contrast to a breeding strategy based on introgression of R genes, the incorporation of prrs would have the potential to confer resistance to a wide range of pathogens. Moreover, given the conserved and essential nature of pamps like ef-tu, mutations are less frequent since they are more likely to penalize pathogen fitness. Thus, it may be less likely for pathogens to evade recognition by prrs through pamp mutations than through effectors mutations34.
The overexpression of a heterologous pamp receptor to confer pti, combined with introgressed quantitative resistance, presumably having different mechanisms of pathogen suppression, may prove more durable resistance than using either approach alone33. This strategy may constitute an important element toward an integrated control of bw in potato.
3.2 Bacterial colonization is restricted in potato lines combining quantitative resistance with atefr effects
In order to gain a much deeper insight into the bacterial infection progress in atefr lines, we inoculated plants with luminescent and fluorescent R. solanacearum reporter strains, and periodically evaluated their colonization patterns. While gfp-labelled reporter strains allowed the detection of bacteria at the single-cell level48, luminescent reporters are more sensitive, allowing a non-destructive in vivo imaging, and quantification of the emitted luminescence that could be correlated with bacterial loads in infected tissues43.
Transgenic plants Iporá atefr 3 and clone 09509.6 atefr 34 were selected for bacterial colonization assays, since these genotypes showed contrasting responses compared to non-transformed genotypes under growth-chamber conditions (data not shown).
Plants inoculated with the UY031 pps-lux were evaluated 3, 4, 5, 6 and 10 days after inoculation (Figure 3). The pathogen was observed for the first time at 5 dai in 7 out of 12 of Iporá wild-type plants, and 6 out of 12 of Iporá atefr 3 plants (Figures 3A, 3B). At this point, luminescent bacteria were not detected in any of the plants derived from clone 09509.6. Only 6 days after inoculation, the pathogen bacteria appeared for the first time in 1 out of 12 plants of the non-transformed clone 09509.6. Plant infection advanced rapidly, and at 10 dai, bacteria had already colonized 9 of these plants, with an extended median colonization pattern (Figure 3C).
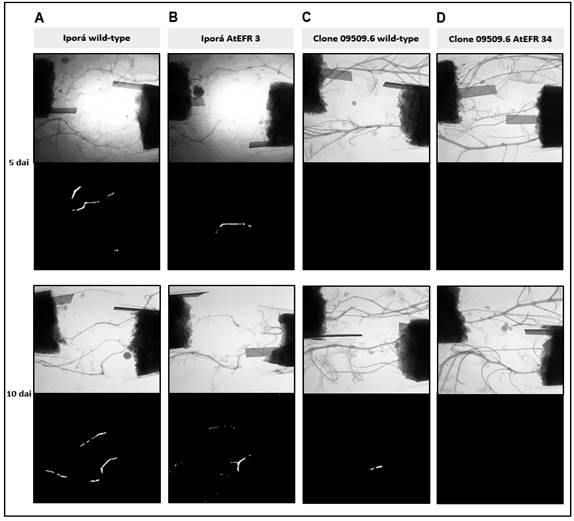
Figure 3: Representative X-ray and luminescence images of potato plants Iporá wild-type (A), Iporá atefr 3 (B), clone 09509.6 wild-type (C), and clone 09509.6 atefr 34 (D) soil inoculated with Ralstonia solanacearum strain UY031 pps-lux. Bacterial colonization was evaluated 3, 4, 5, 6 and 10 dai. In this figure images are presented at 5 dai (the first time R. solanacearum was observed in plants of the susceptible potato cultivar Iporá wild-type and the transgenic line Iporá atefr 3), and 10 dai (the first time it was seen in the partial resistant potato breeding line clone 09509.6 wild-type). Within each observation time, upper images represent X-ray photography, and lower images represent luminescence photography, where luminescent bacteria (R. solanacearum strain UY031 pps-lux) is displayed in white.
However, in clone 09509.6 atefr 34, only 2 out of 12 plants began to show some bacteria on their stems at 10 dai, denoting the strong effect of atefr in this genetic background (Figure 3D). In contrast, 11 out of 12 of the Iporá wild-type plants, and 10 out of 12 of Iporá atefr 3 plants showed a large spread of pathogen through the stem at that time.
In clone 09509.6, the effect of the atefr gene seems to be determinant on delaying the appearance of R. solanacearum, and hindering development of its colonization through the stem. Whereas in Iporá background, the existing difference is not so noticeable between wild-type and transformed plants.
3.3 BW resistance in atefr potato lines is correlated with restricted bacterial colonization at stem-base level
In inoculation assays with the fluorescent reporter strain UY031 pps-gfp, no infection was observed at 2 dai, by microscopic evaluation neither in stems nor in roots (Figure 4 and 5). Only 7 dai, roots from all genotypes were already colonized, but at this point differences in stems colonization between Iporá and clone 09509.6 plants were observed. None of the clone 09506.9-derived plants, neither wild type nor atefr 34, showed fluorescent bacteria at 7 dai (Figures 5A, 5B).
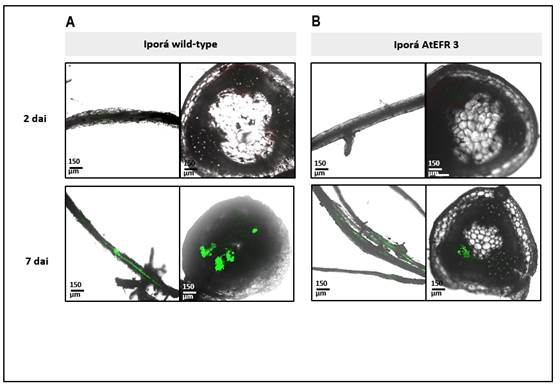
Figure 4: Representative confocal fluorescence micrographs of roots and stem cross-sections of potato plants soil inoculated with Ralstonia solanacearum strain UY031 pps-gfp. Bacterial colonization was evaluated 2 and 7 dai, in the susceptible potato cultivar Iporá wild-type (A), and Iporá atefr 3 (B). Fluorescent bacteria (R. solanacearum strain UY031 pps-gfp) is displayed in green.
At 14 dai, Iporá-derived plants were dead. At this time, bacteria were displayed with a high load and wide distribution in the basal stem of clone 09509.6 wild-type (Figure 5A). Interestingly, fluorescent bacteria were not detected in none of the plants of clone 09509.6 atefr 34 (Figure 5B).
Taking together, atefr transgenic plants show more resistance in part because they are more effective at preventing pathogen multiplication at the stem base, thus hindering its spread to the aerial part, which explains the delayed onset of wilting symptoms in our trial.
In a similar study, it was also shown that efr-based resistance in tomato is due to restricted bacterial colonization from lower stem17. Interestingly, this capability to restrict bacterial colonization by limiting dissemination along the stem was observed in non-transgenic resistant potato32 and tomato plants49)(50.
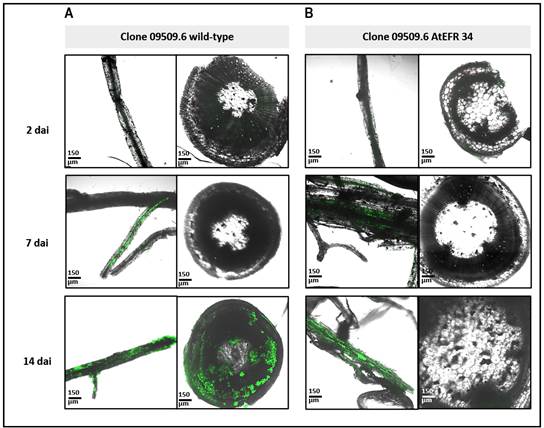
Figure 5: Representative confocal fluorescence micrographs of roots and stem cross-sections of potato plants soil inoculated with Ralstonia solanacearum strain UY031 pps-gfp. Bacterial colonization was evaluated 2 and 7 dai, in the partial resistant potato breeding line clone 09509.6 wild-type (A), and clone 09509.6 atefr 34 (B). Fluorescent bacteria (R. solanacearum strain UY031 pps-gfp) is displayed in green.
4. Conclusions
In this study, we proved that atefr-transformed potato lines display higher bw resistance levels compared to the non-transformed genotypes. The atefr effect was verified for the first time in conditions resembling those in the field. These results highlight the potential of atefr potato plants as a promising commercial crop alternative method. Furthermore, the combination of a pamp receptor reinforcing pti, with introgressed quantitative resistance, turned out to be a valuable strategy, showing better results than using each approach separately.
In addition, by comparing pathogen colonization patterns among the genotypes, we conclude that resistance improvement of atefr genotypes could be attributed to a more effective restriction of the pathogen at stem base, hindering its spread to the aerial part of the plant, which explains the reduced and delayed onset of wilting symptoms. Wholly preventing the spread of the bacteria through the tubers continues to be a goal for improvement.
In prospect, it would be very useful to perform a massive transcriptomic analysis (rna-Seq) in order to identify genes associated with the response mediated by the efr/ef-tu recognition, as well as the effect of minor resistance genes introduced by conventional breeding.