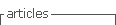1. Introduction
Cyanobacteria, one of the most diverse phototrophic bacterial phyla found in a wide array of habitats, are adapted to a range of environmental conditions. Besides, some of them are capable of fixing atmospheric nitrogen (N) that makes them unique because of their carbon (C) and N autotrophy.
Cyanobacteria play an important role in building soil fertility in rice fields1. Also, several studies underline the role of cyanobacteria as useful biological agents in remediation and amelioration of soil environments2. “Cyanobacterisation”, the use of cyanobacteria as inoculants, has been extensively studied2)(3)(4. These reports show that heterocystous cyanobacteria contribute better to the soil N economy than their unicellular counterparts. Also, indigenous cyanobacterial isolates may be better adapted to local conditions and more suitable for choosing them as potential inoculants5. Cyanobacteria contribution to N in Asian rice paddies is well known. Yet, there is less knowledge about it in rice paddies with a different management or other geographical regions. Rice paddies in Uruguay had a three-year pasture or fallow history, and are dry-seeded with flooding at tillering. The abundance of cyanobacteria in these rice fields is lower than those reported in other ricefields6. Therefore, the inoculation with native cyanobacteria isolates could be a possible N supplement in these ecosystems5. On this regard, cyanobacteria diversity, eco-physiology and potential N fixing activity is relevant for rice paddy sustainability.
Traditionally, the identification of cyanobacteria was based on their morphological characteristics. Their morphological diversity divides the group into five subsections7. The first three subsections comprise unicellular cyanobacteria and filamentous cyanobacteria that do not form heterocytes8, subsection IV (Nostocales) holds filamentous heterocyst-forming cyanobacteria without true branching9, and subsection V (Stigonematales) comprises filamentous heterocyst-forming cyanobacteria with true branching8. Cyanobacteria belonging to subsections IV and V have vegetative cells that can differentiate into morphologically and ultrastructurally distinct cells. One type of differentiated cells is called heterocyst, and has the ability to fix N under aerobic conditions. The other one is called akinete and is a type of resisting cell to thrive under environmental stresses10.
Morphological description and genetic information are not taken together for cyanobacterial identification. Isolates sequences deposited in databases lack a morphological description11. In consequence, a review of the classification of these microorganisms was pointed as necessary7. The nucleic acid-based analyses are the most accepted tool for assessing microbial diversity12. The molecular marker for bacteria is the 16S rRNA gene, because of its universality and conservation and the presence of hypervariable regions13. The resolving power of the 16S rRNA gene is at the species level or above it8. For this reason, it is advisable to carry out a phylogenetic analysis including other molecular markers, as the hetR gene, essential for the differentiation of the heterocyst14, the nifH gene, that encodes for the Fe protein of the nitrogenase15, the rbcL gene, that codifies the large subunit of the Ribulose 1, 5-bi- phosphate carboxylase / oxygenase14, among others.
However, the separate analysis of each of the attributes is not enough to classify them and a polyphasic approach is more appropriate and reliable16. This approach involves the analysis of morphological, molecular, biochemical and physiological characters.
Our aim was to taxonomically classify ten indigenous filamentous heterocystous N-fixing cyanobacteria isolated from a Uruguayan ricefield according to their morphology and phylogeny, and characterize them according to their growth rates and N-related enzymes activities. We wished to find some well-characterized cyanobacteria isolates that could be recommended to be evaluated as rice biofertilizers. A potential biofertilizer candidate should be a well characterized strain, whose presence in soil can be monitored. A high growth rate concomitant with photosynthetic activity may contribute to inoculant formulation and improve its growth and soil colonization. The extent to which cyanobacteria can contribute to the N requirements of crops depends on their N-fixation rate, that is regulated in part for their acclimation to use other forms of N as ammonium (NH4 +) and nitrate (NO3 -). Enzymatic activities related to these latter forms of N were evaluated.
2. Materials and Methods
2.1 Cell growth, culture conditions and growth rates
Ten heterocyst-forming cyanobacteria isolates, hereafter referred as Su1, Su5, Su8, Su10, Su11, Su16, Su37, Su40, Su45, and Su54, were isolated from a Uruguayan rice field17 and kept lyophilized until their use. Cultures were grown in Erlenmeyer flasks under constant agitation in BG11° medium18 at pH 7.6 and buffered with 10 mM HEPES. Cultures were always kept at 28 °C ± 2 °C under white light supplied by fluorescent lamps, providing a photosynthetic photon flux density of 50 mE m-2 s-1 (16:8 light:dark period).
Samples of cyanobacteria grown in BG11° liquid media were taken every two days during ten days to determine growth rates using Optical Density at 750 nm as a pigment independent measurement with a UV-1201V spectrophotometer (Shimadzu).
All measurements for the rest of the determinations were done at the exponential phase of each culture, unless it is indicated.
2.2 Morphological characterization
Features such as vegetative cell shape, heterocyst shape, number and position, and akinete or hormogonia presence were checked with a Nikon Labophot-2 microscope. These data along with the keys from the Bergey’s Manual of Determinative Bacteriology19 were used to assign genera to the isolates. For the observation of akinetes and hormogonia, cultures were left in the same medium during a month without agitation. Illustrations are based on photographs taken with an optic microscope and were edited in Adobe Illustrator.
2.3 Molecular and phylogenetic analyses
Genomic DNA (gDNA) was extracted according to Zhou and others20. First, a 5 mL sample of each cyanobacterium culture was centrifuged at 6,000 rpm for 10 min, previous homogenization with a Potter pestle. Then, a volume of 10 mL of extraction buffer (100 mM Tris HCl, 100 mM EDTA, 100 mM NaH2PO4, 1.5 M NaCl, 1% CTAB, pH 8.0) and proteinase K 0.038 mg mL-1 were added to the precipitate and incubated at 37 °C in agitation for 30 min. One mL of 20% SDS was added, incubated at 65 °C in agitation for 2 h and centrifuged at 6,000 rpm for 10 min. Afterwards, an equal volume of 24:1 chloroform:isoamyl alcohol was added to the supernatant, mixed and centrifuged at 6,000 rpm for 20 min. Isopropanol (0.6 volume) was added to the supernatant, mixed, incubated for 1 h at room temperature, to precipitate the gDNA, and centrifuged at 7,000 rpm for 40 min. Later, the precipitate was washed with cold 70% ethanol and centrifuged at 7,000 rpm for 20 min. The precipitate was dried overnight at room temperature, resuspended in sterile miliQ type ultrapure water, and stored at -20 °C.
The purity of gDNA was checked by measuring A260:A280 ratio in a NanoDrop 2000 spectrophotometer (Thermo Scientific). gDNA quality was checked in a 0.8% agarose gel.
Partial 16S rRNA gene sequence was amplified employing a universal primer for Eubacteria 27F1 (5’-AGAGTTTGATCCTGGCTCAG-3’)21 and a specific primer for cyanobacteria 809R (5’-GCTTCGGCACGGCTCGGGTCGATA-3’)22 (amplicon expected size: 800 bp)..
Partial hetR gene region was amplified using specific primers for cyanobacteria HETR-F1 (5’-TATCTGGCTTTTAGYGCCATG-3’) and HETR-R1 (5’-CTTGGTGATATTTATCWGCCC-3’)14 (amplicon expected size: 740 bp).
Both genes reactions were carried out in 25 μl containing 2.5 μl of 10 X Taq buffer, 1 μl of 10 mM of each deoxynucleotide, 0.5 μl of 10 μM of each primer, 0.20 μl of 5 U Taq DNA polymerase and 2 μl of gDNA extract.
For partial 16S rRNA gene, the pcr program was: initial denaturation at 94 °C for 5 min, 30 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for all the isolates, except for Su5 (temperature of annealing: 62 °C) for 1 min and extension at 72 °C for 1 min, and finally, a final extension at 72 °C for 10 min. For partial hetR gene, the pcr was performed under the following program: initial denaturation at 95 °C for 5 min, 35 cycles of denaturation at 95 °C for 30 s, annealing at 58 °C for 30 s and extension at 72 °C for 1 min. A final extension at 72 °C for 10 min was done.
pcr products were forward and reverse sequenced by Macrogen Inc. (Korea). The sequences were edited manually with the software Chromas Lite 2.123. The partial 16S rRNA gene sequences were deposited in the NCBI GenBank database under the accession numbers: MH271072, MH271073, MH271074, MH271075, MH271076, MH271077, MH271078, MH271079, MH271080, MH271081; and the partial hetR gene sequences under the accession numbers: MH424314, MH424315, MH424316, MH424317, MH424318, MH424319, MH424320, MH424321, MH424322, MH424323. Partial 16S rRNA and hetR genes sequences were aligned against the NCBI GenBank database24, and partial 16S rRNA gene sequences were as well aligned against RDP database25. Phylogenetic trees were built with the Neighbor-joining method, the Maximum Parsimony (mp) and the Maximum Likelihood (ml) algorithms in the MEGA program 426. Chlorobium limicola was used as an out-group taxon for the partial 16S rRNA gene phylogenetic tree. Only heterocyst forming cyanobacteria carries hetR gene, so an outgroup was not included in the phylogenetic analysis.
2.4 Photosynthetic oxygen evolution
Photosynthesis was measured as O2 evolution (nmol min-1 μg-1 Chl a) for 6 min with a Clark-type O2 electrode (Hansatech Instruments Ltd.) as described in Irisarri and others6.. Aliquots of 2 mL of cell suspensions were placed in a cuvette at 27 °C and illuminated with a quantum flux density of 400 mE m-2 s-1.
Chlorophyll a (Chl a) was quantified according to Wellburn27. Briefly, 1 mL of culture was centrifuged at 12,000 rpm during 5 min, previous homogenization with a Potter pestle. The pellet was resuspended in absolute ethanol and incubated overnight at 4 °C. After centrifugation, the supernatant absorbance was read at 665 nm.
2.5 Nitrogenase activity
Gas chromatographic quantification of ethylene form (acetylene reduction activity, ara) was utilized as an index of N fixation. Commercially available standard ethylene was used for quantification. ara was performed in 25 mL aliquots of cell suspensions placed in 60 mL vials incubated with acetylene for 4 h. Ethylene concentrations were determined in a Shimadzu HP 5890 gas chromatograph equipped with a Porapack N column28. Nitrogenase activities were expressed as U mg-1 total proteins-1, where U was μmol ethylene min-1. For total soluble protein quantification, cultures were centrifuged (6,000 g for 10 min) and precipitates were resuspended in an extraction buffer (100 mM Tris; 50 mM EDTA; 100 mM NaCl; 1% PMSF (phenylmethylsulfonyl fluoride), pH 8.0). The protein extract was quantified according to Lowry and others29, and bovine serum albumin was used as standard. To perform ara assay, we selected four isolates, Su5, Su8, Su10 and Su16, due to their morphological differences.
2.6 Nitrate reductase activity
Nitrate reductase (nr) activity was determined as described by Herrero and others30, based on the ability of this enzyme to reduce NO3 - to nitrite (NO2). The isolates were previously cultured for five days in a BG11 medium with 20 mM KNO3. nr activities were expressed as U mg-1 total proteins-1, where U was μmol NO2 - min-1.
2.7 Glutamine synthetase activity
Glutamine Synthetase (gs) activity was performed as described by Dobrogosz31. The gs transferase activities were determined measuring γ-GlutamilHidroxamate (γ-GH) with glutamine as substrate in the presence of ADP and arsenate. gs activities were expressed as U mg-1 total proteins-1, where U was μmolγ-GH min-1.
2.8 Statistical analysis
All experiments described were conducted independently at least twice to confirm the reproducibility of the results. Growth rate, O2 evolution, nitrogenase, nr and gs activities were analyzed through analysis of variance (anova) and tested for differences between isolates using a post-hoc Tukey´s HSD test (p<0.05) in R3.1:3 software using the “agricolae” package32.
3. Results
3.1 Morphological features
The preliminary selection of the studied cyanobacterial isolates was based on their differential colony growth on solid medium (an example in Figures 1B and 2B). Two main types of colonies were observed, gelatinous colonies with different colors (cyan, green or brown) and others velvet-like colonies. The microscopical analysis showed that all of them belong to the order Nostocales (unbranched filaments exhibiting cell differentiation into vegetative cells, heterocytes and akinetes). Within the order, the isolates were divided into two different morphotypes, Nostoc and Calothrix morphotypes (Figures 1 and 2). These in turn matched with the two types of colonies observed. Most of them had simple isopolar filaments without branching, belonging to the Nostocaceae, Su1, Su5, Su8, Su11, Su16, Su40 and Su54, and their genus can be ascribed to Nostoc according to their morphotype (Figure 1A). On the other hand, isolates Su16, Su37 and Su45 belong to the family Rivulariaceae, as they had whip-like tapering filaments, and to the genus Calothrix (Calothrix morphotype, Figure 2). Almost all isolates formed straight filaments, except Su40 and Su54, that had slightly coiled filaments. Heterocytes and akinetes were always found solitary but not only in a particular position within the filament. The information provided by all these features supported the heterogeneity of the isolates.
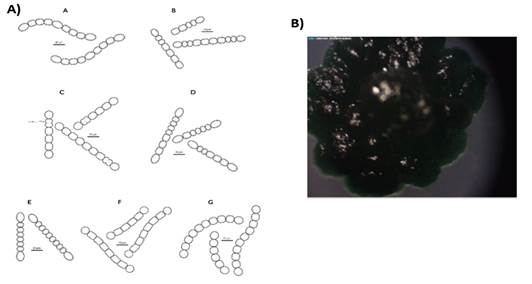
Figure 1: A) Diagrammatic representation of the morphological features of isolates ascribed as Nostoc. A. Su 1. Straight and heteropolar sheathed filament with barrel-shaped cells. Basal, intercalary and oval shaped heterocyst. B. Su 5, straight and heteropolar sheathed filament with cylindrical cells. Single, basal and oval heterocyst. C. Su 8, straight and heteropolar sheathed filament with barrel-shaped cells. Spherical, basal and intercalary heterocyst. D. Su 10, straight and heteropolar sheathed filament with cylindrical cells. Basal, single, intercalary and oval-shaped heterocyst. E. Su 11, straight and heteropolar sheathed filament with cylindrical cells. Basal, single and oval heterocyst. F. Su 40, straight and heteropolar sheathed filament with barreled cells. Heterocyst were spherical, solitary and basal. G. Su 54, straight and heteropolar sheathed filament with cylindrical cells. Heterocyst were basal, solitary and spherical. B) Typical colony grown in BG11o medium.
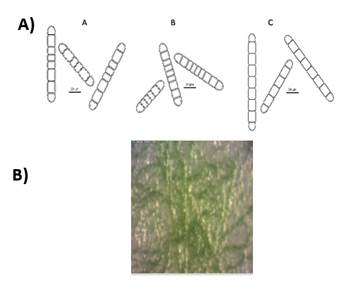
Figure 2: A) Diagrammatic representation of the morphological features of isolates ascribed as Calothrix. A. Su 16, straight, heteropolar sheathed filament with conical end and cylindrical cells. Spherical, basal and single heterocyst. B. Su 37, straight and heteropolar sheathed filament with barrel-shaped cells. Filaments showed conical ends and, in some cases, a false branch. Heterocyst were basal and cylindrical and flattened in one side. C. Su 45, straight, heteropolar sheathed filament with cylindrical cells and conical end. Ovoid heterocysts were basal and solitary. B) Typical colony grown in BG11o medium.
3.2 Phylogenetic analyses
The BLAST of the partial 16S rRNA gene (800 bp fragment) against NCBI GenBank and RDP databases showed that Su1, Su10, Su40, Su16 and Su45 had a high similarity (>97%) with Nostoc elongense, Nostoc piscinale, Nostoc calcicola, Calothrix membranacea and Calothrix parietina, respectively (Table S1, in supplementary material). Su8, Su11 and Su54 showed a high similarity (>99%) with Nostoc sp., Su5 showed a low similarity with Nostoc piscinale (88.5%), and Su37 matched to an uncultured bacterium (Table S1, in Supplementary material).
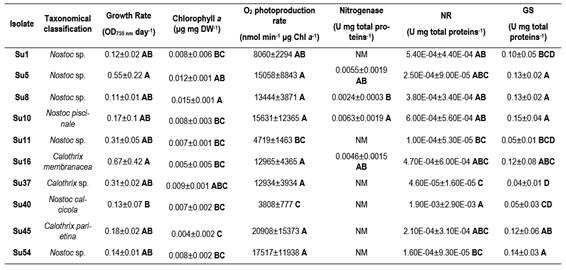
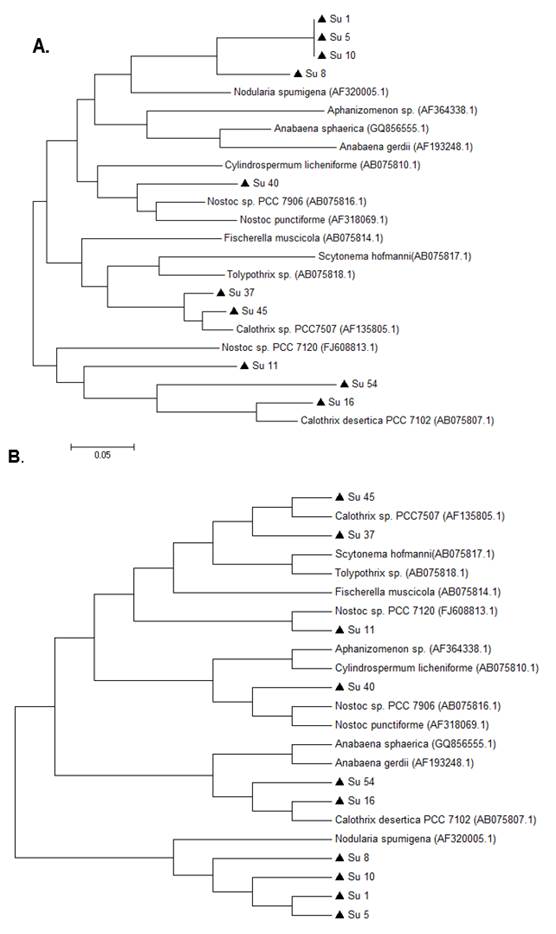
In the 16S rRNA phylogenetic tree, most of the isolates clustered together with the genus Nostoc, except Su45 and Su16, which clustered with Calothrix. The isolate Su37 did not cluster with any of the reference strains. Su5 and Su10 clustered together, like Su54 and Su11 (Figure 3).
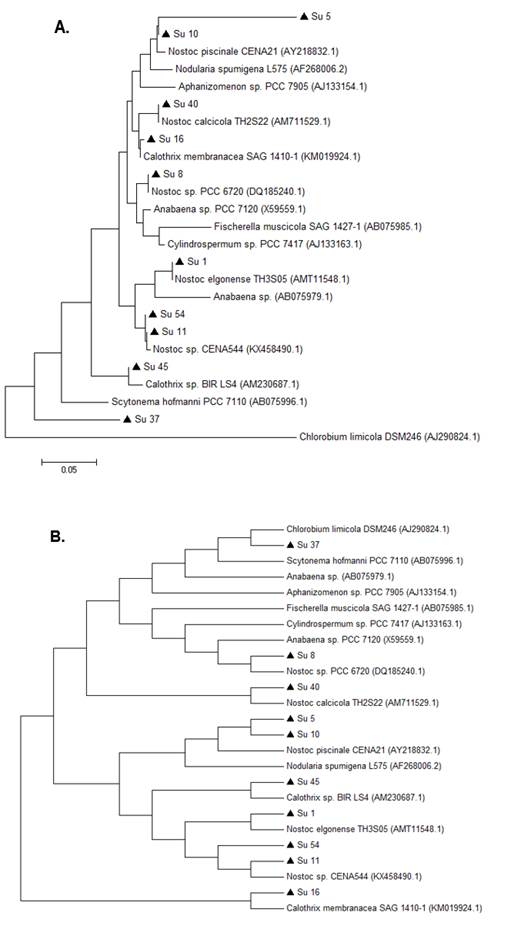
On the other hand, the comparison of the edited sequences of the partial hetR gene of the isolates studied with the NCBI GenBank database did not show any homology equal or greater than 94%, except for Su37 and Su45, that presented a percentage of similarity with the hetR gene of Calothrix sp. (95% and 96%, respectively, Table 2, in supplementary material). The hetR gene BLAST output showed that Su1, Su5, Su8, Su10, Su11 and Su40 shared a low similarity (between 82 and 88 %) with hetR sequences that belong to Nostoc genus, and that Su16 and Su54 shared a low similarity (92% and 80%, respectively) with hetR sequences that belong to Calothrix desertica (Table 2; in Supplementary material).
In the hetR phylogenetic tree, Su1, Su5, Su8 and Su10 clustered together, Su40 and Su11 with two different hetR gene sequences that belong to Nostoc genus, Su37 and Su45 with Calothrix sp, and Su16 and Su54 with Calothrix desertica (Figure 4). Nostoc and Calothrix did not separate into different clades in both phylogenetic trees (Figures 3 and 4).
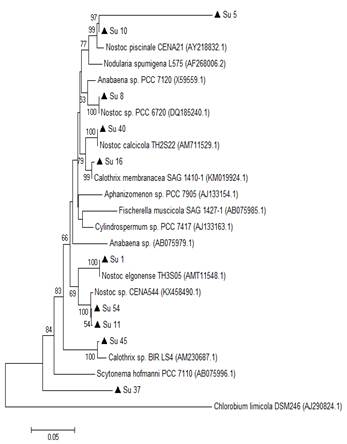
Figure 3: Phylogenetic relationships of the ten isolates (black triangle) based on partial 16S rRNA gene sequences. The tree was built by the Neighbor-joining clustering method. Numbers near nodes indicate bootstrap values >50%. Chlorobium limicola was the outgroup used.

Figure 4: Phylogenetic relationships of the ten isolates (black triangle) based on partial hetR gene sequences. The tree was built by the Neighbor-joining clustering method. Numbers near nodes indicate bootstrap values >50%.
It is worth noticing that the same types of clusterization in the phylogenetic trees were obtained when the Neighbor-joining method, the mp or the ml algorithm were used (Figure S1 and S2; in Supplementary material).
3.3 Generation time and specific growth rate
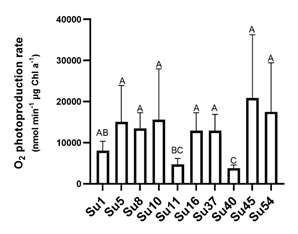
The generation time and specific growth rates were compared among the ten cyanobacterial isolates (Table 1). All isolates had a generation time higher than 35 h with the growth conditions used in this work. The growth rate presented a high standard deviation and only Su5 and Su16 exhibited higher growth rate (0.55 ± 0.22 d-1 and 0.67 ± 0.42 d-1, respectively) than Su40 (0.13 ± 0.07 d-1). The growth rate of the rest of the isolates could not be differentiated.
Table 1: Growth rates of the ten cyanobacterial isolates.
| Isolate | Growth Rate (OD750 nm day-1) |
|---|---|
| Su1 | 0.12±0.02 AB |
| Su5 | 0.55±0.22 A |
| Su8 | 0.11±0.01 AB |
| Su10 | 0.17±0.1 AB |
| Su11 | 0.31±0.05 AB |
| Su16 | 0.67±0.42 A |
| Su37 | 0.31±0.02 AB |
| Su40 | 0.13±0.07 B |
| Su45 | 0.18±0.02 AB |
| Su54 | 0.14±0.01 AB |
Same capital letter showed no statistical difference (p<0.05).
3.4 Oxygen photoevolution
The isolates photosynthetic activities were estimated by their O2 production. The rates of O2 photoevolution were similar for all isolates, although Su45 exhibited the maximum rate, it was only significantly different to Su11 and Su40 (Figure 5).
3.5 Nitrogen related enzymes
Nitrogenase activities were measured with the acetylene reduction assay for the isolates Su5, Su8, Su10 (Nostoc-morphotype) and Su16 (Calothrix-morphotype). Su10 recorded the highest average nitrogenase activity, followed by Su5 and Su16, although the activity of these three isolates was not significantly different (Figure 6C). Only Su8 exhibited a lower nitrogenase activity than Su10 (Figure 6C).
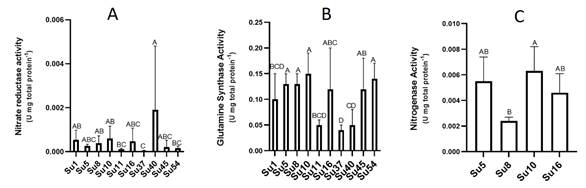
Figure 6: A) Nitrate reductase (nr) activity of the ten cyanobacteria isolates B) Glutamine synthase (gs) activity of the ten cyanobacteria isolates C) Nitrogenase activity of four of the cyanobacteria isolates. Bars values are the means of four replicates and error bars represent standard errors. Significant differences among isolates are indicated by capital letters, according to ANOVA-Tukey`s HSD (p<0.05).
In spite of the low nr activities determined (Figure 6A), Su40 showed the highest nr activity, while Su37 the lowest. Most of the isolates did not show significant differences among them.
Regarding gs activities, isolates Su5, Su8, Su10, Su16, Su45 and Su54 presented the highest gs activity and Su37 the lowest (Figure 6B).
All the isolates`characteristics are summarized in Supplementary Table 3.
4. Discussion
In the present study, we aimed to characterize ten heterocyst-forming cyanobacteria by analyzing their morphological and molecular features (polyphasic approach). We also assessed some of their physiological-biochemical traits in order to select potential inoculants for their use in ricefields.
4.1 Morphological and molecular analyses
The ten heterocyst-forming isolates selected for this study presented two main different types of colonies (Fig. 1B and Fig. 2B), although the colonies were all different in color or other characteristic. Morphological diversity may be explained by cyanobacteria great acclimatizing ability in response to growth conditions and may lead to their misidentification33. In concert with this, some authors claimed that more than 50% of the strains in culture collections are misidentified34. In this work the ten studied isolates were all grown under the same conditions and maintained their morphological characteristics. The morphological characteristics of the isolates permit to ascribe them to genera Nostoc and Calothrix. Other reliable methods, such as 16S rRNA gene and hetR gene sequencing were used in order to confirm the morphological characterization. By definition, two isolates are members of the same species when they share at least 97% of similarity of the 16S rRNA gene sequence35. However, Stackebrandt and Ebers36 recommended to set a stricter range to describe the uniqueness of a new bacterial isolate. These authors claimed that this percentage of similarity should be, at least, 98.7%36. This last criteriont allowed to identify four isolates at species level, Su10 as Nostoc piscinale (99% of similarity), Su 40 as Nostoc calcicola (100% of similarity), Su16 as Calothrix membranacea (99.7% of similarity), and Su 45 as Calothrix parietina (98.7% of similarity). Considering other authors37, isolate Su1 could have been described as Nostoc elgonense (97% of similarity). For this particular case and in order to improve the phylogenetical characterization of cyanobacterial isolates that were not assigned at species level, other gene markers such as Internally Transcribed Spacer (its) between the 16S and 23S rRNA sequences could be used in next studies. This marker has been recently used for the study of intra-species Mycrocistis diversity by Next-Generation Sequencing38.
Isolates Su5, Su8, Su11 and Su54 were assigned to Nostoc genus according to the morphological characterization and 16S rRNA phylogenetic analysis. However, this was not achieved for isolate Su37 although the hetR phylogenetic analysis along with the morphological characterization were congruent for this isolate (Calothrix genus). There was another case where the output of the different approaches was not similar. For instance, the hetR phylogenetic analysis of Su54 was not congruent with the morphological and 16S rRNA analysis. This may be explained as the database for hetR gene sequences is smaller compared to the 16S rRNA database39. The 16S rRNA gene database is larger and updated providing a more robust phylogenetic reconstruction and better taxonomic identification. Besides that, the 16S rRNA gene does not suffer from horizontal gene transfer and it is more conserved in function and in structure than the genes that code for proteins as hetR gene, as reviewed by Barker and others40. In general, cyanobacteria classification is troublesome due to the presence of sequences of cyanobacterial species in databases without morphological description, and classification of species based solely on morphology without any genetic information41. In this study, almost in all cases, the correlation between morphological characterization and phylogenetic analysis was good.
Phylogenetic analyses of both genes showed that the studied isolates ascribed as Nostoc and Calothrix intermingled in one clade. Other studies have reported these results before14)(16. This can be explained as heterocyst-bearing cyanobacteria (sections IV and V) form a monophyletic cluster despise their cellular differentiation.
4.2 Potential inoculant evaluation
Heterocyst-forming cyanobacteria have been widely used as biofertilizers in agriculture, especially in Asian paddy rice fields2. It is well known that indigenous strains or isolates, better adapted to local conditions, are more suitable to be applied in the field42. In terms of inoculant formulation, there is a stricter legal regulation in Uruguay than in Asian countries. For instance, in India, a common practice is to use an unknown formulation in the field. In our country, every inoculant has to be thoroughly identified and tested before its use in the field43. For this reason, our aim focused on the characterization and testing of some physiological and biochemical characteristics to evaluate their growth in vitro. Hence, providing preliminary information to select cyanobacterial isolates to be used as inoculants for rice.
In rice fields, Nostoc sp. is one of the most abundant heterocyst-forming cyanobacteria and mostly used as inoculants for rice, wheat, corn and other crops42)(44)(45. Calothrix sp. has also been reported in rice fields and assayed as biofertilizers46)(47)(48.
The first parameters to consider when selecting bacterial inoculants are their generation time and specific growth rate. These provides data to determine how fast the isolates can grow or their adaptation to in vitro conditions, etc. The studied isolates did not show significant differences in their growth rates or generation time. However, Su16 and Su54 had the highest growth rates. Su40 showed one of the lowest generation times and a low rate of photosynthesis. For this reason, isolate Su40 may not be a good choice to select as inoculant. On the other hand, the growth rate was not related to any genus of cyanobacteria, as one Nostoc sp. and one Calothrix sp. isolates presented the higher values. Isolates within the same genus presented the highest and lowest growth rates.
Heterocyst-forming cyanobacteria perform photosynthesis in vegetative cells and N fixation in heterocyst where nitrogenase is expressed. In order to assess their capability to fix N2, some isolates were incubated with acetylene. Our results indicate that Su10, Su5 and Su16 could be considered potential biofertilizers. Yet caution has to be taken as the acetylene reduction assay to estimate nitrogenase activity has many drawbacks49. It was not possible to observe a higher heterocyst frequency in the isolates with the highest nitrogenase activity (data not shown) as was reported before50. Hrckova and others51 reported higher nitrogenase activity for strains of Tolypothrix (similar to Calothrix) than for strains of Nostoc. These authors explained that Nostoc strains produce large amounts of mucus and create thick envelopes for cell protection52, reducing diffusion and light penetration and limiting nitrogenase activity. In our study, this difference was not observed.
N fixation and heterocyst differentiation are highly regulated responding to deprivation of combined N53. But cyanobacteria can also use other N sources, as NO3 - or NH4 +. In case of NH4 +, cyanobacteria preferred N source, it can be directly assimilated whether it comes from biological N fixation (bnf) or from its uptake from the soil solution53. gs is one of the enzymes involved in NH4 + incorporation into C backbones and so its role is relevant. In our study, all isolates had a low nr activity. This can be explained maybe due to a short time left to induce nr activity30. Su37 had the lowest nr activity and also the lowest gs activity. Su5, Su10 and Su16 showed high nr and gs activities, suggesting a high capacity to assimilate NH4+. This also matches with the fact that this isolate had a high photosynthetic activity, and probably a higher amount of C backbones for N assimilation.
Although some cyanobacterial strains from rice fields were shown to release small quantities of NH4 + during their active growth phase54, in general, most of the fixed products are made available mainly through microbial mineralization55. Strains of cyanobacteria that release NH4 + continuously were suggested as a solution to control the N availability to rice plants56. Thus, isolates with a high gs activity would not be good candidates as biofertilizers, due to their incapability to release NH4 + to the environment. In this case, Su11, Su37 and Su40 showed lower gs activities, although they did not show the highest values regarding the the other physiological and biochemical parameters determined in this work. Thus, further studies are necessary in this regard.
5. Conclusions
Morphological and molecular characterization let us conclude that the ten isolates corresponded to different strains.
Likewise, isolates Su5 (Nostoc sp.), Su10 (Nostoc piscinale) and Su16 (Calotrhix membranace) may be the best candidates to be used as rice biofertilizers. They depicted high-growth rates, photosynthetic and nitrogenase activities. However, their gs activity was high indicating they may not release NH4 +.