Services on Demand
Journal
Article
Related links
Share
Odontoestomatología
Print version ISSN 0797-0374On-line version ISSN 1688-9339
Odontoestomatología vol.24 no.39 Montevideo June 2022 Epub Aug 01, 2022
https://doi.org/10.22592/ode2022n39e315
Update
Molecular bases of benign odontogenic tumors: a review of the literature in the context of the latest classification of the World Health Organization
1 Área de Patología Molecular Estomatológica, Facultad de Odontología, Universidad de la República (UDELAR), Montevideo, Uruguay
Odontogenic tumors (OTs) are a heterogeneous group of lesions that range from hamartomas to benign or malignant neoplasms. The present study aimed to review the literature regarding the molecular and genetic aspects from some benign OTs. Based on the review of the included studies, it is supported that genetic background may play an important role in the etiology and pathogenesis of OTs. However, the body of evidence on the subject still does not allow to conclude the definitive mechanisms involved in the development and progression of OTs. It is suggested the development of further molecular and genetic studies evaluating significant samples of OTs to establish its etiopathogenesis, facilitate its diagnostic process and enrich the therapeutic approaches.
Keywords: odontogenic tumors; molecular pathology
Los tumores odontogénicos (TOs) son un grupo heterogéneo de lesiones que incluyen hamartomas hasta neoplasias benignas o malignas. El presente estudio tuvo como objetivo revisar la literatura sobre los aspectos moleculares y genéticos de algunos TOs benignos. A partir de los estudios revisados, se apoya la idea de que las causas genéticas pueden tener un papel importante en la etiología y en la patogénesis de algunos TOs. Sin embargo, las investigaciones sobre el tema aún no permiten concluir de manera sólida los mecanismos definitivos involucrados en el desarrollo y en la progresión de los TOs. Se sugiere el desarrollo de más estudios moleculares y genéticos que evalúen muestras significativas de TOs para establecer su etiopatogenia, facilitar su proceso diagnóstico y enriquecer su abordaje terapéutico.
Palabras clave: Tumores odontogénicos; Patología molecular
Os tumores odontogênicos (TOs) são um grupo heterogêneo de lesões que variam desde hamartomas até neoplasias benignas ou malignas. O presente estudo teve como objetivo revisar a literatura sobre os aspectos moleculares e genéticos de alguns TOs benignos. A partir dos estudos revisados, apoia-se que as causas genéticas podem desempenhar um papel importante na etiologia e patogênese de alguns TOs. No entanto, as pesquisas sobre o assunto ainda não permitem uma conclusão sólida sobre os mecanismos definitivos envolvidos no desenvolvimento e progressão dos TOs. Sugere-se o desenvolvimento de mais estudos moleculares e genéticos para avaliar amostras significativas de TOs para estabelecer sua etiopatogenia, facilitar seu processo diagnóstico e enriquecer sua abordagem terapêutica.
Palavras-chave: Tumores odontogênicos; Patologia molecular
Introduction
Odontogenic tumors (OTs) are rare pathological entities arising from odontogenic tissues or their remnants in the gnathic bones.1 OTs are a heterogeneous group of pathologies ranging from hamartomatous lesions to benign and malignant neoplasms, with highly heterogeneous clinical and histopathological behaviors.2,3 The related epidemiological data vary according to the geographic region and the population studied. Its reported prevalence is less than 1% among all tumors affecting the head and neck region. The classification of OTs is a hotly debated topic in science. The World Health Organization (WHO) published its classification in previous editions (1992 and 2005). The classification was revised and updated in 2017.2 Throughout the study of OTs, attempts have been made to simplify the classification of this heterogeneous group of lesions based mainly on their histogenesis.3
There have been advances in immunohistochemical studies, molecular biology, genetic knowledge, and the clinical and epidemiological follow-up of OTs. Therefore, in 2017 the WHO modified its classification to discard clinically irrelevant lesion subtypes.4 This publication distinguishes between benign and malignant OTs. According to their origin, benign OTs were subdivided into epithelial, mixed (epithelial and mesenchymal), and mesenchymal, significantly more frequent than malignant odontogenic lesions.4,5 Malignant OTs are classified into carcinomas, sarcomas and odontogenic carcinosarcoma. These arise de novo or from a previous benign odontogenic lesion.4 This classification is dynamic as new entities are included and others are eliminated as our knowledge about OTs advances (Table 1).
New studies have researched the molecular and genetic basis of OTs, enriching the knowledge about its potential etiopathogenesis.6 Frequent specific mutations have been reported in specific odontogenic lesions that would have clinical significance.7-9 However, the etiopathogenesis of this heterogeneous group of lesions is not completely established. In this context, this narrative literature review aims to determine the clinicopathological aspects and review the main findings of the molecular pathology of OTs in the context of WHO’s latest classification published in 2017.
Methods
The following electronic databases were consulted: PubMed, Web of Science, Embase, and Scopus. We considered the most significant studies in the scientific literature on this subject. They were selected considering studies previously published in the literature on OTs until August 2020 (no pre-set start date). The following keywords were used for the search: “odontogenic tumors” in the first stage; “odontogenic tumors” AND “review” in the second stage; and “odontogenic tumors” AND “molecular” in the final stage. In addition, terms specific to each type of OT were included in the search. Finally, we reviewed the secondary references identified in the literature of the primary publications found in the databases. The articles were selected by convenience, randomly by title and abstract, and organized by topic according to OT type. The criteria for choosing the articles were based on OT molecular research. The articles were included in the relevant groups according to OT type for further analysis and extraction of the primary data.
Literature review and discussion
Main molecular and genetic aspects of odontogenic tumors
Various studies suggest that molecular and genetic changes might be involved in the development and progression of OTs.3,6 Changes in oncogenes and tumor suppressor genes may participate in the etiopathogenesis of OTs.10 This section addresses some classical and recent evidence on the molecular and genetic aspects of OTs, highlighting the involvement of oncogenes, tumor suppressor genes, DNA repair genes, growth factors, cell cycle regulators, apoptotic factors, regulators of tooth development, cell adhesion molecules, extracellular matrix degradation proteins, angiogenic factors, and osteolytic cytokines.
Oncogenes promote neoplastic transformation by activating gene amplification, mutation, or translocation. The products of these genes function as growth factors and growth factor receptors, such as epidermal growth factor receptor (EGFR), fibroblast growth factor (FGF), thrombocyte-derived growth factor (PDGF), HER-2, and signal transducers (Ras).6,10 These factors are involved in proliferation and differentiation events. In OTs, products encoded by the Ras oncogene, p21Ras, for example, are involved in the transduction of external growth factor-induced stimuli and may be more highly expressed in OTs than in normal odontogenic tissue. In addition, genetically modified mice bearing the Myc and H-Ras oncogenes—potentially involved in cell proliferation events—had a high incidence of OTs.11,12 The above results may suggest a potential connection between oncogenes and the etiopathogenesis/biological behavior of OTs.
Tumor suppressor genes reduce the likelihood that specific cells will become cancerous. When these genes are inactivated, usually by a genetic mutation, their mechanism of action is deregulated, and tumors may develop. Examples of tumor suppressor genes involved in the development of neoplasms are p53 (the most frequently altered gene in tumors and with a major role in response to genomic damage), retinoblastoma (Rb), WT-1, adenomatous polyposis coli (APC), and patched (PTC).6 Previous studies demonstrated increased p53 immunoexpression in benign and malignant OTs.13-16). The APC gene and its product have been linked to the Wnt signaling pathway and inhibition of cell proliferation. In this context, one study demonstrated that APC immunohistochemical reactivity was lower in ameloblastoma cases than in tooth germs.17
DNA repair genes, such as MSH2 and MHL1, act by adjusting potential errors related to DNA replication and repair. In the immunohistochemical study conducted by Castrilli et al.,18 hMSH2 and hMLH1 proteins were expressed in ameloblastoma neoplastic cells. This suggests that the tumor’s development/progression does not depend on defects in the DNA maintenance system.
Growth factors play a role in controlling proliferation and differentiation. In this sense, dysfunctions in growth factors can lead to pathological conditions. According to previous studies, some growth factors potentially involved in the origin of OTs are transforming growth factor alpha (TGF-α), EGFR, and TGF-β.12. Kumamoto et al.51 ran immunohistochemistry tests and demonstrated positive expression of hepatocyte growth factor (HGF) and TGF-β and its receptors in tooth germs and OTs of epithelial origin. Therefore, these growth factors might have potential action in epithelial cells. Furthermore, the authors detected altered expressions of these growth factors in epithelial OTs. Therefore, they also conclude their potential influence on the differentiation of odontogenic neoplastic epithelial cells. Yamada et al.20) demonstrated that TGF-β1 growth factor-β1 and interleukin-1α produced by odontogenic cysts and OTs induce osteoclastogenesis by inducing the expression of receptor activator ligand for nuclear factor kappa-B (RANKL) in the stroma. This evidence may reinforce the potential involvement of various growth factors in the biological behavior of OTs.
Cell cycle regulators are related to cell proliferation events and may be involved in tumor development when deregulated. OT proliferation has been evidenced by numerous proliferation and cell cycle markers, such as proliferating cell nuclear antigen (PCNA), Ki-67, and DNA topoisomerase IIα.21-24 Shahela et al.24 evaluated the cellular kinetics of dentigerous cysts, radicular cysts, odontogenic keratocysts, and ameloblastomas to study the differences between these odontogenic lesions through the immunohistochemical expression of the cell cycle antigen PCNA. The study showed that the odontogenic keratocyst presented the highest protein expression values among the odontogenic cysts evaluated. Furthermore, ameloblastoma presented the highest PCNA expression values. This proves the potential connection between cell cycle markers and the growth patterns of odontogenic lesions.
Apoptotic factors are also studied in the molecular pathology of odontogenic neoplasms. Apoptosis may be regulated by tumor necrosis factor (TNF), Bcl-2, and the inhibitor of apoptosis protein (IAP). Apoptotic cells have been observed in some OTs, suggesting that apoptosis is involved in oncogenesis and cell differentiation of the odontogenic epithelium.25 Mascitti et al.26 evaluated the expression of p73 (apoptosis-related) protein and TNF-related apoptosis-inducing ligand (TRAIL) in odontogenic tumors and cysts. They showed that imbalances in apoptotic pathways might participate role in OT development. Also, p53 may act in apoptotic induction in cases of irreversible genetic damage. Tenório et al.27 studied the expression of p53 and other apoptosis and cell cycle regulatory proteins (Bcl-2, Bax) in odontogenic epithelial lesions. They found various expression patterns in different lesions and their potential connection with their biological behaviors.
Various regulators of tooth development, which determine positioning and shape (Msx-1 and -2, Dlx-2, Barx-2), and tooth morphogenesis and cytodifferentiation (Sonic hedgehog , bone morphogenetic proteins , Wnt), may also be involved in OT pathogenesis.6,28 In the study conducted by Zhang et al.,29 the SHH signaling pathway, which has a role in tooth development, and its receptor PTC, SMO, and transcription factor GLI1, were predominantly located in the epithelial component of various types of OTs. This suggests their potential involvement in the proliferation of epithelial components in OTs. Even in this context, Santos et al.30 suggested the involvement of Regγ, Wnt-1, and β-catenin proteins in the pathogenesis of benign epithelial odontogenic lesions. Finally, other factors potentially involved in OT development and progression are cell adhesion molecules, extracellular matrix degradation proteins, angiogenic factors, and osteolytic cytokines.6 Carreón-Burciaga et al.31 showed differences in the expression of cell adhesion molecules E-cadherin and Syndecan-1 in different types of ameloblastomas associated with tumor size and recurrence. This suggests a potential connection between the expression of these cell adhesion molecules and the potential aggressiveness of these tumors. Kumamoto and Ooya32 demonstrated that BMP and associated molecules might play a role in the cytodifferentiation of the normal and neoplastic odontogenic epithelium through epithelium/mesenchyme interaction. Kumamoto and Ooya33 demonstrated that vascular endothelial growth factor (VEGF) can be considered a major molecule in the regulation of angiogenesis in epithelial OTs and that its overexpression can be associated with neoplastic or malignant changes in odontogenic epithelial cells. Sá et al.34 suggested the potential involvement of IL-α, TNF-α, and IL-10 in the pathogenesis of odontogenic tumors and cysts. Osteolytic factors have a higher immunohistochemical expression in tumors with more aggressive biological behavior. Andrade et al.35 demonstrated the expression of bone metabolism regulators RANK, RANKL, and osteoprotegerin (OPG) in different OTs. This suggests their potential association with local bone resorption events in these tumors. In brief, extensive scientific evidence shows various molecular, genetic and epigenetic aspects potentially involved in the etiopathogenesis of OTs. Below we present the results of molecular analyses in some specific types of benign OTs.
Ameloblastoma
Recently, researchers have detected associations of mutations responsible for the dysregulation of various cellular functions in the mitogen-activated protein kinase (MAPK) pathway. This is the case especially in B-Raf oncogene (BRAF), in approximately 60% of cases of ameloblastomas; in the BRAF activating protein called RAS, which includes KRAS, NRAS, and HRAS; and in fibroblast growth factor receptor 2 (FGRF2), which activates the MAPK signaling pathway (Figure 1).36-38 Other studies have also addressed the potential associations between gene changes to repair DNA damage and the development of ameloblastomas.39 Sweeney et al. 8 identified BRAF mutations that resulted in a valine to glutamic acid substitution at codon 600 in 46% of cases in an ameloblastoma sample. Surprisingly, SMO mutations exhibited a marked preponderance in maxillary ameloblastomas, whereas BRAF mutations exhibited a reverse pattern, with higher frequency in mandibular cases. Various studies have detected this mutation in ameloblastoma cases. Fregnani et al.40 studied if BRAF-V600E expression would be associated with a more aggressive clinical presentation or molecular pattern in ameloblastomas. A large immunohistochemical panel (cytokeratins , EGFR, PTHrP, syndecan-1, Ki-67, p53, and BRAF-V600E) was performed on 73 cases of ameloblastomas, and several clinicopathological parameters were collected to assess potential associations. This study found BRAF-V600E mutations in 34 cases (46%). This was statistically associated with CK8, CK16, PTHrP, and p53 expressions, with recurrences, multilocular radiographic appearance, and cortical bone disruption. In addition, univariate analyses showed that BRAF-positive cases were associated with lower disease-free survival rates. Therefore, the study suggests associations between BRAF-V600E positivity and parameters related to greater clinical aggressiveness in ameloblastomas. Lapthanasupkul et al.41 also evaluated the frequency of BRAF-V600E mutations in conventional and unicystic ameloblastomas in a group of Thai patients. Cases of ameloblastoma with clinicopathologic information were selected for immunohistochemical evaluation of anti-BRAF V600E antibody. The authors demonstrated BRAF-V600E positivity in 72.5% of the conventional ameloblastomas evaluated and found no association with any of the clinicopathologic parameters evaluated. Unicystic ameloblastoma samples showed a 95.5% positivity rate. Given the high BRAF-V600E positivity of the cases considered, the authors suggest the possibility of future BRAF-targeted therapies to manage mutated ameloblastomas. Other studies have also shown associations between BRAF-V600E-positive ameloblastomas with a higher disease-free survival rate.36 This shows the importance of further studies with a significant number of ameloblastoma cases to better elucidate the lesion’s molecular aspects.
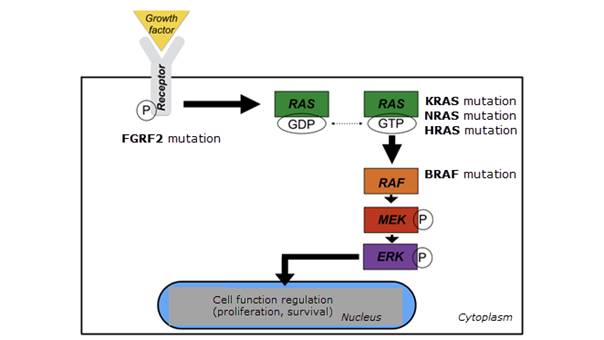
Figure 1: Diagram of the mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) pathway of some of the mutations observed in the ameloblastoma.
In a study on molecular events in ameloblastomas, Amaral-Silva et al.42 evaluated the expression pattern of proteins responsible for identifying and correcting DNA damage (hMSH2, hMSH3, and hMSH6) in developing human odontogenic tissues, ameloblastomas, and ameloblastic carcinomas. The authors sought to determine whether the expression of these proteins could be associated with the prognosis of ameloblastomas. For that purpose, samples were organized to evaluate protein expression, and 73 other cases of ameloblastoma with clinicopathological information and BRAF-V600E mutations were simultaneously evaluated with tissue microarray. hMSH2 and hMSH6 expression was significantly lower in ameloblastomas than in developing human tooth samples. In addition, hMSH2 and hMSH3 expression was significantly associated with BRAF-V600E mutation. Cases with simultaneous expression of the studied proteins were associated with recurrence but without an association with predicted disease-free survival. These results show that the proteins evaluated were less expressed in ameloblastomas and were associated with recurrence but without the ability to predict disease-free survival. This study allows us to infer a connection between changes in the proteins responsible for identifying and correcting DNA damage and ameloblastoma development. This reinforces the need for further studies on the subject and shows the controversies around the definition of the molecular aspects of OTs. In this context, another study by Bologna-Molina et al.43 analyzed the immunohistochemical expression of DNA repair proteins hMLH1 and hMSH2, and Ki-67 in ameloblastomas and tooth germs. The study included 40 conventional ameloblastomas, 40 unicystic ameloblastomas, and 5 tooth germs. Unicystic ameloblastomas showed higher hMLH1 and hMSH2 expression than conventional ones. Ki-67 expression was inversely proportional to the expression of DNA repair proteins. In addition, the expression of HMLH1, hMSH2, and Ki-67 was significantly higher in tooth germs than in the ameloblastomas in the study. The authors concluded that the expression of DNA repair proteins and Ki-67 could be correlated with the biological behavior of ameloblastomas and the physiological mechanisms of tooth germs.
Given the importance of angiogenesis for tumor maintenance, growth, and spread, Montezuma et al.44 studied if the expression of cyclooxygenase-2 (COX-2)—a molecule associated with an increased number of blood vessels in pathological processes—is related to microvascular density and tumor aggressiveness in ameloblastomas. The authors used immunohistochemical techniques with COX-2 and CD34 in 63 ameloblastoma cases with clinicopathologic information collected from patients’ medical records. BRAF-V600E expression was obtained from a previous study conducted by the same research group. Of the cases evaluated, 28 (44.4%) were positive for COX-2 with a mean microvascular density of 2.2 vessels/mm2. Specific statistical analyses were conducted. Then, the authors observed associations between COX-2 immunohistochemical expression and recurrence and BRAF-V600E expression. In addition, a lower microvascular density was associated with conservative therapy in the ameloblastoma cases evaluated. Finally, COX-2 expression was significantly associated with a lower five-year disease-free survival rate, but without an association with higher microvascular density. These molecular studies on ameloblastomas suggest various molecular approaches potentially involved in their etiopathogenesis.
Assessing genetic changes in ameloblastomas has also yielded significant results related to a potential role in the pathogenesis of this type of OT. Various molecular and genetic dysregulations can affect the development and oncogenic transformation of the odontogenic epithelium into ameloblastoma.45 Numerous signal transduction pathways are closely associated with the occurrence, development, and prognosis of ameloblastoma. MAPK transduces intracellular signals and is reported to contribute significantly to the pathogenesis of ameloblastomas. It also mediates BRAF V600E activation.46 In the research of genetic alterations involved in the etiopathogenesis of ameloblastomas, some studies have also evaluated the influence of the phosphatase and tensin homologous gene (PTEN), which controls cell migration and proliferation and monitors Akt level, in ameloblastoma pathogenesis. Narayan et al.47 studied the genetic changes in exon 5 of the PTEN gene in patients with ameloblastoma by extracting genetic material from paraffin-embedded biopsies of 20 conventional ameloblastomas and 10 tooth germs. Of the 20 ameloblastomas, 5 showed genetic alterations. Of these, 3 (15%) showed silent mutation, 1 (5%), change in amino acid sequence from valine to glutamic acid, and 1 (5%), mRNA decay. This study showed a 25% somatic mutation of frequency in exonic region 5 of the PTEN gene, which may indicate a potential role in the pathogenesis of ameloblastoma. Therefore, genetic pathways might be involved in the pathogenesis of ameloblastomas.
Adenomatoid odontogenic tumor
In Reichart et al.’s review48 on the immunohistochemical profile of adenomatoid OTs, the authors state that this type of lesion has been extensively studied, as reported in several studies. Regarding the etiology of adenomatoid OTs, the authors say that the lesion seems to derive from the dental lamina or its remnants. Furthermore, the immunohistochemical pattern update performed in this review seemed sufficient to explain that adenomatoid OTs may be more of a hamartomatous nature than neoplasms per se. This conclusion was based on the following main points listed in the study:
(1) Cytokeratin profiles and some integrin types are invasion related in ameloblastoma but not in adenomatoid OTs;
(2) High expression of p53 protein in ameloblastoma, but not in adenomatoid OTs;
(3) Preferential or much increased levels of the proliferative marker MDM2 in ameloblastoma are not as readily demonstrable in adenomatoid OTs;
(4) Lower labeling indices of cell proliferation and anti-apoptosis markers Ki-67 and Bcl-2 in adenomatoid OTs;
(5) Metallothionein levels in ameloblastoma are significantly higher than in adenomatoid OTs, conferring lower recurrence risk and recurrence potential; (6) Lower levels of matrix metalloproteinase levels in adenomatoid OTs correspond to lesser degrees of local aggressiveness.
Bello et al.49 studied the immunohistochemical expression pattern of FAK, paxillin, and PI3K in ameloblastomas and adenomatoid OTs to assess the connection between these major proteins and the development of neoplasms. Immunohistochemical techniques were used to study FAK, paxillin, and PI3K in 45 ameloblastomas, 7 adenomatoid OTs, and 2 developing human teeth. The authors observed weak FAK expression in adenomatoid OTs and variable expression in ameloblastomas (from weak to strong). The expression of paxillin and PI3K was relatively similar in the two tumor types evaluated. In tooth germs, FAK and paxillin expression was strong in all the components of the enamel organ, whereas PI3K expression was strong in the inner epithelium of the enamel organ. The authors concluded that the immunohistochemical expression of these proteins in tooth germs would suggest a significant role in odontogenesis. As for the tumors, the study found weak FAK expression in adenomatoid OTs compared to strong expression in some ameloblastomas. This suggests that this molecular difference might be involved in the patterns of biological behavior of these two types of lesions: the more indolent behavior of adenomatoid OTs compared to the more aggressive profile of ameloblastoma. This relationship may be justified by the association between FAK and tumor aggressiveness, local invasion, and metastasis in various studies of malignant neoplasms.50,51 However, this study evaluated immunohistochemical expression patterns in specific lesion samples. Further studies assessing other samples of this type of OT are needed to draw more conclusions about these results.
As noted, the pathogenesis of adenomatoid OTs is not fully established. Some studies point to a potential genetic alteration involved in the development of adenomatoid OTs, especially in the KRAS gene.52 The protein encoded by the KRAS gene is related to the EGFR transduction cascade and its mutation has been demonstrated in various neoplasms.53 A study by Bologna-Molina et al.54 studied the frequency of the KRAS mutation and its association with the presence of the MAPK/ERK signaling pathway in adenomatoid OTs. Genetic material was collected from nine cases of adenomatoid OTs. In addition, genetic mutations related to 50 genes associated with neoplasm development were evaluated with next-generation sequencing in one case. Mutations in the RAS family (Luminex assay) were considered in the other eight cases. Finally, immunohistochemical tests for KRAS, CRAF, BRAF, EGFR, ERK, MEK, and BRAFV600E were performed in all cases. The study showed a KRAS gene mutation at codon 12 but not in the stromal tissue of the lesions. KRAS G12V and KRAS G12R mutations were detected in two and four cases, respectively. Regarding the immunohistochemical tests, all cases were positive for EGFR, KRAS, BRAF, and CRAF, one case was negative for ERK, and another case was negative for MEK and ERK. The study concluded that the activated MAPK/ERK signaling pathway accompanied by the KRAS gene mutation at codon 12 might be a pathogenic factor involved in adenomatoid OT development. Several studies evaluate the mechanisms potentially involved in the etiopathogenesis of adenomatoid OTs. However, more scientific evidence is needed to understand the molecular aspects of this lesion.
Calcifying epithelial odontogenic tumor
Some studies have evaluated the participation of matrix proteins expressed during tooth enamel formation (enamelin, amelogenin, amelotin, ameloblastin, and ameloblast-associated odontogenic protein) in odontogenic neoplasms. The expression of these proteins may be significant markers for understanding the functional differences between different types of OTs. Crivellini et al.55 evaluated the immunohistochemical expression of these proteins in adenomatoid OTs, calcifying epithelial OTs, odontomas, ameloblastomas, calcifying odontogenic cysts, ameloblastic fibromas, myxomas, odontogenic fibromas, and reduced enamel epithelium. Considering the heterogeneity of OTs and the pluripotent nature of the odontogenic tissues involved, the results of this study on calcifying epithelial OTs showed immunohistochemical expression of amyloid substances and part of neoplastic cells for odontogenic ameloblast-associated protein. Immunohistochemical expression of amelotin was restricted to the cell cytoplasm of some neoplastic cells, and the ameloblastin marker was positive in one of the tumors and in some neoplastic cells. Finally, amelogenin expression was observed around some calcified structures. The immunohistochemical expression patterns of the odontogenic ameloblast-associated protein are addressed in other studies. Crivellini et al.55 suggest that the expression of this protein in calcifying epithelial OTs is due to the stimulation of mineral formation that occurs in this type of OT. In addition, the authors state that calcifying epithelial OTs probably originate from ameloblastic cell lines because the affinity of the odontogenic ameloblast-associated protein for the mineralized structure only occurs in odontogenic epithelia with this cell type. The authors draw several conclusions on the histopathogenetic theories based on the expression of the proteins evaluated. For instance, they report that the epithelium of calcifying epithelial OTs, adenomatoid OTs, and odontomas showed specialized secretory activity inherent to ameloblasts and different from that observed in ameloblastomas, ameloblastic fibromas, and odontogenic fibromas. In this same scenario, Perdigão et al.’s study56 evaluated the ameloblastin gene in different types of OTs. The protein expressed by this gene is related to the differentiation and proliferation of odontogenic cells. The authors sequenced the coding regions of this gene in one case of calcifying epithelial TO, two cases of calcifying odontogenic cysts and one case of ameloblastic fibroma. They found that the genetic sequencing was altered only in the calcifying epithelial OT. Therefore, the authors suggest that this genetic alteration may be relevant in the pathogenesis of this type of OT.
Sousa et al.57 evaluated mutations in oncogenes and tumor suppressor genes in calcifying epithelial OTs. In this study, 50 genes commonly mutated in neoplasms were sequenced in 9 calcifying epithelial OTs with next-generation sequencing. The study showed that only one case evaluated harbored mutations in the PTEN and CDKN2A suppressor genes and in the JAK3 and MET oncogenes. As this occurred in a single tumor, the authors suggest that these changes are probably unrelated to mutations in calcifying epithelial OTs. Peacock et al. also evaluated the genetic profile of calcifying epithelial OTs.58 They detected changes in the PTCH1 gene with immunohistochemical techniques and genetic material sequencing. PTH1 is a tumor suppressor gene whose mutation is associated with some syndromes and the sporadic development of some neoplasms. The authors suggest that this genetic alteration may be involved in the pathogenesis of calcifying epithelial OT.
Odontogenic myxoma
Santos et al.59 evaluated potential genetic mutations in odontogenic myxomas by evaluating 50 genes commonly mutated in neoplasms. The authors evaluated nine odontogenic myxoma samples with next-generation sequencing. They showed that there were no cases with pathogenic genetic mutations. Variations in KDR, TP53, PIK3CA, KIT, and JAK3 were detected. Therefore, the authors concluded that odontogenic myxomas do not appear to have genetic mutations that commonly occur in malignant neoplasms or may appear in a low proportion of cases. Odontogenic myxomas may present mutations in other genes that were not evaluated in this study or the variations presented may have a specific pathogenic role in this type of lesion.
Odontogenic myxomas can present myofibroblastic differentiation and myxoid areas similar to intraosseous myofibromas. Siqueira et al.60 compared cases of odontogenic myxomas with myofibromas. Recent studies have demonstrated mutations of the PDGFRB gene in single and multiple cases of myofibromas. The authors sequenced exons 12 and 14 of the PDGFRB gene in 15 cases of odontogenic myxomas. The study detected a typical pattern of the sequenced gene in all odontogenic myxoma samples. This suggests that the PDGFRB gene does not play a role in the pathogenesis of this type of OT. Furthermore, the authors conclude that this may be a differential marker between odontogenic myxomas and myofibromas.
Odontogenic myxomas can present local aggressiveness and a high recurrence rate. Osteoclastogenesis is important in terms of neoplasm growth. In this context, González-Galván et al.’s study61 evaluated the immunohistochemical expression pattern of the proteins related to OPG, RANK, and RANK-L osteolytic events in odontogenic myxomas. The study included 18 cases of odontogenic myxomas—4 small and 14 large—and 18 dental follicles. The authors demonstrated significant differences in RANK expression in odontogenic myxomas compared to dental follicles, and in the comparison between small and large lesions. The RANK-L and OPG markers showed no statistically significant differences. However, the authors mention a pattern of higher marker expression in large odontogenic myxomas. They concluded that RANK/RANK-L/OPG activation would participate in the mechanism of osteolytic regulation and tumor growth in cases of odontogenic myxomas. In this context, changes in this signaling pathway could be involved in the more aggressive mechanism of bone invasion in some cases of odontogenic myxomas. Bologna-Molina et al.62 evaluated the immunohistochemical expression pattern of VEGF and ORM-1 protein—whose concentrations are related to homeostasis disorders and tumor growth—in 33 cases of odontogenic myxomas and in 3 tooth germs. Immunohistochemical tests were performed to evaluate proteins in tumor cells, endothelial cells, and the extracellular matrix. Over half of the cases did not present VEGF immunoexpression in the extracellular matrix, and ORM-1 expression was strong in endotheliocytes and tumor cells and positive in the extracellular matrix of odontogenic myxomas, varying from moderate to strong in all cases. With these results, the authors suggest that VEGF and ORM-1 expression may be associated with angiogenesis and structural viscosity that influence tumor growth in odontogenic myxomas.
Primordial odontogenic tumor
Mosqueda-Taylor et al.63 described six cases of primordial odontogenic tumors. Immunohistochemical study of the lesions showed that mesenchymal tumor cells labeled positively for vimentin and negatively for α-smooth muscle actin, desmin, S100, and CD34. Ki-67 expression was low, and the epithelial component was strongly positive for AE1/3 and CKs 5 and 14 immunoexpression. Azzi et al.64 conducted a systematic literature review to update the data on primary OTs. In this work, 18 cases of primordial OTs were identified. The mean age observed was 11.58 years, with cases ranging from 2 to 19 years and a slight preference for the male sex (61.11%). Asymptomatic enlargement of the posterior mandible was the most common clinical feature. The most common radiographic finding was well-defined uni- or multilocular radiolucent lesions, and most cases were surgically enucleated. In this context, Bologna-Molina et al.65 performed an extensive immunohistochemical test on four cases of primordial OTs to study their potential histogenesis and biological behavior. The study showed that the epithelial component lining the surface of the tumors was positive for CKs 14 and 19. The expression of amelogenin, glut-1, MOC-31, caveolin-1, PITX2, p53, Bax, Bcl-2, survivin, and PTEN showed focal variations. The mesenchymal component was positive for vimentin, syndecan-1, PITX2, CD105, CD34, cyclin D1, Bax, Bcl-2, survivin, and p53. PTEN and CD90 were moderately positive. BRAF V600E and calretinin were negative in all four samples. Ki-67 and MCM-7 were expressed in less than 5% of tumor cells. The study concluded that primordial OTs are benign tumors whose histogenesis involves epithelial and mesenchymal activities.
To investigate the pathogenesis of primordial OTs, Mikami et al.66 evaluated six cases with DNA and transcriptome analysis with next-generation sequencing. In addition, immunohistochemical tests of amelogenin, ameloblastin, and dentin sialophosphoprotein were performed. The evaluation of the results showed no genetic mutations in any gene related to odontogenesis or neoplasm development. Genes encoding enamel (Amelx, Ambn, and Enam) and dentin (Col1a1, Dspp, Nes and Dmp1) proteins were expressed. Genes associated with dentinogenesis (Bglap, Ibsp, and Nfic) were weakly expressed or negative, suggesting inhibition of dentin formation in primordial OTs. Expressions of amelogenin, ameloblastin, and dentin sialophosphoprotein were detected. This study concluded that the pathogenesis of primordial OTs can be considered genetically different from the other types of OTs. It is suggested that the inhibition of enamel and dentin formation may be related to defects in the dentinogenesis process.
Conclusions
Several controversial points related to the classification, etiopathogenesis, and molecular aspects of OTs are constantly discussed in the scientific arena. This literature review lists various molecular, genetic, and epigenetic alterations potentially involved in the development and progression of odontogenic lesions. However, based on the range of results of the multiple studies presented, determining the etiology of OTs remains a challenge. This variability of results may be justified by the characteristic heterogeneity observed in the group of OTs. Therefore, further molecular and genetic studies evaluating significant samples of OTs are suggested to consolidate the knowledge on the etiopathogenesis of odontogenic lesions. A better understanding of the etiology and pathogenesis of odontogenic neoplasms may facilitate diagnosis and enrich therapeutic management. Additionally, a better understanding of the underlying molecular mechanisms will help predict the course of odontogenic tumors. It will also help develop new therapeutic applications for odontogenic tumors, such as molecular targeted therapy and patient-tailored therapy.
REFERENCES
1. Bilodeau EA, Seethala RR. Update on Odontogenic Tumors: Proceedings of the North American Head and Neck Pathology Society. Head Neck Pathol. 2019;13(3):457-465. [ Links ]
2. Wright JM, Soluk Tekkesin M. Odontogenic tumors: where are we in 2017 ?. J Istanb Univ Fac Dent. 2017;51(3 Suppl 1):S10-S30. [ Links ]
3. Sandoval-Basilio J, González-González R, Bologna-Molina R, et al. Epigenetic mechanisms in odontogenic tumors: A literature review. Arch Oral Biol. 2018;87:211-217. [ Links ]
4. El-Naggar 2017 El-Naggar AK, Chan JKC, Grandis JR, Takata T, Slootweg P, eds. WHO classification of head and neck tumours. 4th ed. Lyon: IARC Press; 2017. [ Links ]
5. Barrios-Garay K, Agudelo-Sánchez L, Aguirre-Urizar J, Gay-Escoda C. Analyses of odontogenic tumours: the most recent classification proposed by the World Health Organization. Med Oral Patol Oral Cir Bucal. 2020;23751. [ Links ]
6. Kumamoto H. Molecular pathology of odontogenic tumors. J Oral Pathol Med. 2006;35(2):65-74. [ Links ]
7. Yukimori A, Oikawa Y, Morita KI, et al. Genetic basis of calcifying cystic odontogenic tumors. PLoS One. 2017;12(6):e0180224. [ Links ]
8. Sweeney RT, McClary AC, Myers BR, et al. Identification of recurrent SMO and BRAF mutations in ameloblastomas. Nat Genet. 2014;46(7):722-725. [ Links ]
9. Kurppa KJ, Catón J, Morgan PR, et al. High frequency of BRAF V600E mutations in ameloblastoma. J Pathol. 2014;232(5):492-498. [ Links ]
10. Garg K, Chandra S, Raj V, Fareed W, Zafar M. Molecular and genetic aspects of odontogenic tumors: a review. Iran J Basic Med Sci. 2015;18(6):529-536. [ Links ]
11. Gibson CW, Lally E, Herold RC, Decker S, Brinster RL, Sandgren EP. Odontogenic tumors in mice carrying albumin-myc and albumin-rats transgenes. Calcif Tissue Int. 1992;51(2):162-167. [ Links ]
12. Dodds AP, Cannon RE, Suggs CA, Wright JT. mRNA expression and phenotype of odontogenic tumours in the v-Ha-ras transgenic mouse. Arch Oral Biol. 2003;48(12):843-850. [ Links ]
13. Sandra F, Nakamura N, Kanematsu T, Hirata M, Ohishi M. The role of MDM2 in the proliferative activity of ameloblastoma. Oral Oncol. 2002;38(2):153-157. [ Links ]
14. Batista de Paula AM, da Costa Neto JQ, da Silva Gusmão E, Guimarães Santos FB, Gomez RS. Immunolocalization of the p53 protein in a case of ameloblastic fibrosarcoma. J Oral Maxillofac Surg. 2003;61(2):256-258. [ Links ]
15. Kumamoto H, Izutsu T, Ohki K, Takahashi N, Ooya K. p53 gene status and expression of p53, MDM2, and p14 proteins in ameloblastomas. J Oral Pathol Med. 2004;33(5):292-299. [ Links ]
16. Tenório JR, Santana T, Queiroz SI, de Oliveira DH, Queiroz LM. Apoptosis and cell cycle aberrations in epithelial odontogenic lesions: An evidence by the expression of p53, Bcl-2 and Bax. Med Oral Patol Oral Cir Bucal. 2018;23(2):e120-e125. [ Links ]
17. Kumamoto H, Ooya K. Immunohistochemical detection of beta-catenin and adenomatous polyposis coli in ameloblastomas. J Oral Pathol Med. 2005;34(7):401-406. [ Links ]
18. Castrilli G, Piantelli M, Artese L, et al. Expression of hMSH2 and hMLH1 proteins of the human DNA mismatch repair system in ameloblastoma. J Oral Pathol Med. 2001;30(5):305-308. [ Links ]
20. Yamada C, Aikawa T, Okuno E, et al. TGF-ß in jaw tumor fluids induces RANKL expression in stromal fibroblasts. Int J Oncol. 2016;49(2):499-508 [ Links ]
21. Kim J, Yook JI. Immunohistochemical study on proliferating cell nuclear antigen expression in ameloblastomas. Eur J Cancer B Oral Oncol. 1994;30B(2):126-131. [ Links ]
22. Slootweg PJ. p53 protein and Ki-67 reactivity in epithelial odontogenic lesions. An immunohistochemical study. J Oral Pathol Med. 1995;24(9):393-397. [ Links ]
23. Kumamoto H, Kimi K, Ooya K. Detection of cell cycle-related factors in ameloblastomas. J Oral Pathol Med. 2001;30(5):309-315. [ Links ]
24. Shahela T, Aesha S, Ranganathan K, et al. Immunohistochemical expression of PCNA in epithelial linings of selected odontogenic lesions. J Clin Diagn Res. 2013;7(11):2615-2618. [ Links ]
25. Kumamoto H, Ooya K. Immunohistochemical and ultrastructural investigation of apoptotic cell death in granular cell ameloblastoma. J Oral Pathol Med. 2001;30(4):245-250. [ Links ]
26. Mascitti M, Santarelli A, Zizzi A, Procaccini M, Lo Muzio L, Rubini C. Expression of p73 and TRAIL in odontogenic cysts and tumors. J Oral Sci. 2016;58(4):459-464. [ Links ]
27. Tenório JR, Santana T, Queiroz SI, de Oliveira DH, Queiroz LM. Apoptosis and cell cycle aberrations in epithelial odontogenic lesions: An evidence by the expression of p53, Bcl-2 and Bax. Med Oral Patol Oral Cir Bucal. 2018;23(2):e120-e125. [ Links ]
28. Ruhin-Poncet B, Ghoul-Mazgar S, Hotton D, et al. Msx and dlx homeogene expression in epithelial odontogenic tumors. J Histochem Cytochem. 2009;57(1):69-78. [ Links ]
29. Zhang L, Chen XM, Sun ZJ, Bian Z, Fan MW, Chen Z. Epithelial expression of SHH signaling pathway in odontogenic tumors. Oral Oncol. 2006;42(4):398-408. [ Links ]
30. Santos HBP, Medeiros HCM, Mafra RP, Miguel MCC, Galvão HC, de Souza LB. Regulation of Wnt/ß-catenin pathway may be related to Reg? in benign epithelial odontogenic lesions. Oral Surg Oral Med Oral Pathol Oral Radiol. 2019;128(1):43-51. [ Links ]
31. Carreón-Burciaga RG, González-González R, Molina-Frechero N, López-Verdín S, Pereira-Prado V, Bologna-Molina R. Differences in E-Cadherin and Syndecan-1 Expression in Different Types of Ameloblastomas. Anal Cell Pathol (Amst). 2018;2018:9392632. [ Links ]
32. Kumamoto H, Ooya K. Expression of bone morphogenetic proteins and their associated molecules in ameloblastomas and adenomatoid odontogenic tumors. Oral Dis. 2006;12(2):163-170. [ Links ]
33. Kumamoto H, Ohki K, Ooya K. Association between vascular endothelial growth factor (VEGF) expression and tumor angiogenesis in ameloblastomas. J Oral Pathol Med. 2002;31(1):28-34. [ Links ]
34. Sá MC, de Matos FR, Conceição TS, Leitão AC, Freitas RA. Immunoexpression of tumour necrosis factor-a, interleukin-1a and interleukin-10 on odontogenic cysts and tumours. Int Endod J. 2017;50(5):437-445. [ Links ]
35. Andrade FR, Sousa DP, Mendonça EF, Silva TA, Lara VS, Batista AC. Expression of bone resorption regulators (RANK, RANKL, and OPG) in odontogenic tumors. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2008;106(4):548-555. [ Links ]
36. Brown NA, Rolland D, McHugh JB, et al. Activating FGFR2-RAS-BRAF mutations in ameloblastoma. Clin Cancer Res. 2014;20(21):5517-5526. [ Links ]
37. Diniz MG, Gomes CC, Guimarães BV, et al. Assessment of BRAFV600E and SMOF412E mutations in epithelial odontogenic tumours. Tumour Biol. 2015;36(7):5649-5653. [ Links ]
38. Tan S, Pollack JR, Kaplan MJ, Colevas AD, West RB. BRAF inhibitor treatment of primary BRAF-mutant ameloblastoma with pathologic assessment of response. Oral Surg Oral Med Oral Pathol Oral Radiol. 2016;122(1):e5-e7. [ Links ]
39. Toprani SM. DNA damage and repair scenario in ameloblastoma. Oral Oncol. 2020;108:104804. [ Links ]
40. Fregnani ER, Perez DE, Paes de Almeida O, et al. BRAF-V600E expression correlates with ameloblastoma aggressiveness. Histopathology. 2017;70(3):473-484. [ Links ]
41. Lapthanasupkul P, Laosuk T, Ruangvejvorachai P, Aittiwarapoj A, Kitkumthorn N. Frequency of BRAF V600E mutation in a group of Thai patients with ameloblastomas. Oral Surg Oral Med Oral Pathol Oral Radiol. 2020;S2212-4403(20)31034-8. [ Links ]
42. Amaral-Silva GKD, Sánchez-Romero C, Wagner VP, et al. Prognostic significance of hMSH2, hMSH3, and hMSH6 expression in ameloblastoma. Oral Surg Oral Med Oral Pathol Oral Radiol. 2017;124(3):286-295. [ Links ]
43. Bologna-Molina R, Pereira-Prado V, Sánchez-Romero C, et al. Expression of hMLH1 and hMSH2 proteins in ameloblastomas and tooth germs. Med Oral Patol Oral Cir Bucal. 2018;23(2):e126-e131. [ Links ]
44. Montezuma MAP, Fonseca FP, Benites BM, et al. COX-2 as a determinant of lower disease-free survival for patients affected by ameloblastoma. Pathol Res Pract. 2018;214(6):907-913. [ Links ]
45. Nagi R, Sahu S, Rakesh N. Molecular and genetic aspects in the etiopathogenesis of ameloblastoma: An update. J Oral Maxillofac Pathol. 2016;20(3):497-504. [ Links ]
46. You Z, Liu SP, Du J, Wu YH, Zhang SZ. Advancements in MAPK signaling pathways and MAPK-targeted therapies for ameloblastoma: a review. J Oral Path Med. 2019;48(3):201-205. [ Links ]
47. Narayan B, Urs AB, Augustine J, et al. Genetic alteration of Exon 5 of the PTEN gene in Indian patients with ameloblastoma. Oral Surg Oral Med Oral Pathol Oral Radiol. 2019;127(3):225-230. [ Links ]
48. Reichart PA, Philipsen HP, Khongkhunthian P, Sciubba JJ. Immunoprofile of the adenomatoid odontogenic tumor. Oral Dis. 2017;23(6):731-736. [ Links ]
49. Bello IO, Alrabeeah MA, AlFouzan NF, Alabdulaali NA, Nieminen P. FAK, paxillin, and PI3K in ameloblastoma and adenomatoid odontogenic tumor. (published online ahead of print), 2020 Jul 17. Clin Oral Investig. 2020;10.1007/s00784-020-03465-4 [ Links ]
50. McLean GW, Carragher NO, Avizienyte E, Evans J, Brunton VG, Frame MC. The role of focal-adhesion kinase in cancer - a new therapeutic opportunity. Nat Rev Cancer. 2005;5(7):505-515. [ Links ]
51. Deramaudt TB, Dujardin D, Noulet F, et al. Altering FAK-paxillin interactions reduces adhesion, migration and invasion processes. PLoS One. 2014;9(3):e92059. [ Links ]
52. Gomes CC, de Sousa SF, de Menezes GH, et al. Recurrent KRAS G12V pathogenic mutation in adenomatoid odontogenic tumours. Oral Oncol. 2016;56:e3-e5. [ Links ]
53. Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53(4):549-554. [ Links ]
Bologna-Molina R, Ogawa I, Mosqueda-Taylor A, Takata T, Sánchez-Romero C, Villarroel-Dorrego M, Takeda Y, Mikami T. Detection of MAPK/ERK pathway proteins and KRAS mutations in adenomatoid odontogenic tumors. Oral Dis. 2019;25(2):481-487 [ Links ]
55. Crivelini MM, Felipini RC, Miyahara GI, de Sousa SC. Expression of odontogenic ameloblast-associated protein, amelotin, ameloblastin, and amelogenin in odontogenic tumors: immunohistochemical analysis and pathogenetic considerations. J Oral Pathol Med. 2012;41(3):272-280. [ Links ]
56. Perdigão PF, Carvalho VM, DE Marco L, Gomez RS. Mutation of ameloblastin gene in calcifying epithelial odontogenic tumor. Anticancer Res. 2009;29(8):3065-3067. [ Links ]
57. de Sousa SF, Diniz MG, França JA, et al. Cancer genes mutation profiling in calcifying epithelial odontogenic tumour. J Clin Pathol. 2018;71(3):279-283. [ Links ]
58. Peacock ZS, Cox D, Schmidt BL. Involvement of PTCH1 mutations in the calcifying epithelial odontogenic tumor. Oral Oncol. 2010;46(5):387-392. [ Links ]
59. Santos JN, Sousa Neto ES, França JA, et al. Next-generation sequencing of oncogenes and tumor suppressor genes in odontogenic myxomas. J Oral Pathol Med. 2017;46(10):1036-1039. [ Links ]
60. de Siqueira EC, de Sousa SF, Carlos R, et al. Odontogenic myxomas lack PDGFRB mutations reported in myofibromas. J Oral Pathol Med. 2020;49(3):278-283. [ Links ]
61. González-Galván MC, Mosqueda-Taylor A, Bologna-Molina R, Setien-Olarra A, Marichalar-Mendia X, Aguirre-Urizar JM. Evaluation of the osteoclastogenic process associated with RANK / RANK-L / OPG in odontogenic myxomas. Med Oral Patol Oral Cir Bucal. 2018;23(3):e315-e319. [ Links ]
62. Bologna-Molina R, Mosqueda-Taylor A, Domínguez-Malagón H, et al. Immunolocalization of VEGF-A and orosomucoid-1 in odontogenic myxoma. Rom J Morphol Embryol. 2015;56(2):465-473. [ Links ]
63. Mosqueda-Taylor A, Pires FR, Aguirre-Urízar JM, et al. Primordial odontogenic tumour: clinicopathological analysis of six cases of a previously undescribed entity. Histopathology. 2014;65(5):606-612. [ Links ]
64. Azzi L, Tettamanti L, Di Francesco A, et al. Primordial odontogenic tumour: A systematic review of the common but also unusual features of this novel entity. J Stomatol Oral Maxillofac Surg. 2020;S2468-7855(20)30042-2. [ Links ]
65. Bologna-Molina R, Mikami T, Pereira-Prado V, Pires FR, Carlos-Bregni R, Mosqueda-Taylor A. Primordial odontogenic tumor: An immunohistochemical profile. Med Oral Patol Oral Cir Bucal. 2017;22(3):e314-e323. [ Links ]
66. Mikami T, Bologna-Molina R, Mosqueda-Taylor A, et al. Pathogenesis of primordial odontogenic tumour based on tumourigenesis and odontogenesis. Oral Dis. 2018;24(7):1226-1234. [ Links ]
Conflict of interest declaration: The authors have no conflict of interest regarding the publication of this paper.
Authorship contribution and collaboration statement: a) Conception and design of study; b) Acquisition of data; c) Data analysis; d) Discussion of results; e) Drafting of the manuscript; f) Approval of the final version of the manuscript) Silveira FM has contributed in a, b, c, e, f. Pereira-Prado V has contributed in a, c, e, f. Bologna-Molina has contributed in a, c, e, f.










 text in
text in