Services on Demand
Journal
Article
Related links
Share
Odontoestomatología
Print version ISSN 0797-0374On-line version ISSN 1688-9339
Odontoestomatología vol.17 no.25 Montevideo May 2015
Detection and prevalence of periodontal pathogens in a Uruguayan population with chronic periodontitis using conventional methodology and metagenomics
Papone, Virginia*, Verolo, Carolina*, Zaffaroni, Lourdes*, Batlle, Alicia**, Capo, Claudia**, Bueno, Luis**, Gamonal, Jorge***, Silva, Nora***, Soria, Sandra*
* Microbiology Department, School of Dentistry, Universidad de la República. Uruguay. vpaponey@gmail.com
** Cllinic of Periodontics, School of Dentistry, Universidad de la República. Uruguay
*** School of Dentistry, Universidad de Chile. Chile
Abstract
Periodontal diseases are a major problem in human health. Decades of research have shown that the most common disease is chronic periodontitis, characterized by a slow evolution with the formation of periodontal pockets, subsequent alveolar bone resorption, loss and destruction of teeth and bone tissue. While we know the multifactorial origin of the development of periodontitis, the participation of subgingival microbiota is relevant in the etiology of periodontal disease. Some pathogenic bacteria species that have been associated with the development of periodontal disease are Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, Tannerella forsythia, Fusobacterium nucleatum, among others. In this work we studied which of these five species were present in the periodontal pockets of 51 Uruguayan patients with chronic periodontitis. To achieve the results a conventional microbiological technique and metagenomics (multiplex-PCR) were used.
The results of the microbiological conventional technique showed the presence of A. actinomycetemcomitans (33%) and black pigmented anaerobic bacteria (100%) in the samples. From the results obtained in the multiplex-PCR we saw that the most prevalent species were F. nucleatum (100%), T. forsythia (92%) and P. gingivalis (88%). In contrast, lower prevalence species were P. intermedia (39%) and A. actinomycetemcomitans (33%).
Keywords: microbiota, prevalence, periodontal disease, metagenomics, multiplex-PCR.
Received on: 07.01.15 - Accepted on: 26.03.15
Introduction
Oral cavity infections are a serious public health issue worldwide given their high prevalence in the adult and child population. The etiology of these pathologies includes many factors: the host, the environment and infectious factors. These pathologies pose the potential risk of tooth loss caused by chronic periodontitis in children and adults, as well as the possible development of infective endocarditis and even oral and colorectal cancer (1-6). In the case of chronic periodontitis or periodontal disease there is loss of attachment and bone loss (horizontal and vertical alveolar bone). This disease has its highest prevalence in adults (>40 years), although it can appear in people with their primary or permanent teeth. It is a slowly evolving disease with the following symptoms: gingival swelling with edema, increase in gingival volume, round gingival margins, flattened papilla, accumulation of supragingival and subgingival plaque, formation of calculus, greater tooth mobility and exfoliation. All these symptoms can affect a variable number of functional teeth in each individual, with variable progression rates (7, 8). Over 300 bacterial species associated with periodontitis have been isolated in periodontal pockets. However, only a small percentage of them are etiologically relevant, such as the group of Gram-negative anaerobic pigmented bacilli that belong to the genera Porphyromonas, Prevotella, Bacteroides and Fusobacterium, of the Bacteroidaceae family. Additionally, studies conducted to identify species that are potential risk indicators in the development of periodontitis have included: Aggregatibacter actinomycetemcomitans (Aa), Porphyromonas gingivalis (Pg), Prevotella intermedia (Pi), Tannerella forsythia (Tf) and Fusobacterium nucleatum (Fn), among others (7-10).
More studies are conducted globally on the composition of the oral microbiota of periodontopathic patients using different methodologies, as seen in the work of Herrera et al. in Colombia, Spain and Chile. In South America, the F. nucleatum species is the most prevalent one in Chile (90%) (11); in Colombia, however, two species showed an 80% prevalence: P. gingivalis and T. forsythia (12, 13). In studies conducted on Brazilian patients with periodontitis, P. intermediate and F. nucleatum were the periodontal pathogens present, with a detection frequency of 67%. The least prevalent species were P. gingivalis (26.7%), T. forsythia (20%) and A. actinomycetemcomitans (13%) (14).
Patients with oral infections have been monitored in Uruguay since the 1990s (Faculty of Microbiology, School of Dentistry, Universidad de la República). Pathogens are identified using conventional microbiological methodologies, and the results obtained guide the treatment for each patient. We must remember that conventional methods of direct diagnosis have serious limitations, such as: a) transport and conservation of the sample; b) sample processing; d) development time (2–14 days) and the difficulties for culture growth (15, 16). It is essential to use metagenomic methods to reduce the time it takes to obtain results and to increase detection sensitivity and speciation. Metagenomics is an approach to the study of microbial communities, defined as the functional and sequence analysis of collective microbial genomes in a sample of the oral cavity. It can be based on expression or sequencing (17, 18). For example, the multiplex-PCR technique (multiplex polymerase chain reaction) is frequently used in the direct detection of the microbiota metagenome composition at species and/or serotype level, without having to harvest, isolate and morphologically characterize pathogens that grow with difficulty and/or slowly. The molecular technique uses oligonucleotides complementary to regions conserved in the DNA of each species that codifies the small subunit of 16S ribosomal RNA, in this case to detect periodontal pathogens (11, 19-22).
In this study we performed the rapid and simultaneous identification of the microbiota involved in oral processes using multiplex-PCR, jointly with conventional microbiological methodology, on 51 Uruguayan patients attending the Clinic of Periodontics of the School of Dentistry, Universidad de la República.
Methodology
Patient selection
A total of 51 patients who were ≥ 30 years old were selected (23 women and 28 men) presenting symptoms of mild chronic periodontitis (CAL = 3-4 mm) and advanced chronic periodontitis (CAL ≥ 5 mm), bleeding on probing, pathological pockets and bone loss. Periodontal diagnosis was conducted following the parameters of the American Academy of Periodontology (AAP) (23) with at least 12 teeth present excluding the third molar, without prior periodontal treatment; patients free of diabetes, arthritis, ulcerative colitis, HIV, cancer and cardiovascular pathology. The following types of patients were also excluded: with initial periodontitis, (patient with clinical attachment loss of between 1 and 3 mm) pregnant women, and those who had been treated with antibiotics and/or anti-inflammatories two months before the study. All patients were notified of the research and agreed to sign a consent form before a witness, according to the Regulations of the MERCOSUR. The Ethics Committee of the School of Dentistry, Universidad de la República, approved the design of the study following the regulations of the MERCOSUR and the Helsinki Declaration on research involving human subjects.
Samples
In each quadrant 4 areas were selected based on X-rays and clinical study, with pockets ≥ 5.0 mm deep and ≥ 2.0 mm bone loss. Supragingival plaque was removed using sterile gauze trying to avoid bleeding, and the area was dried with sterile cotton rolls. Medium sterile paper points (Nº 25) were placed deeply into the pocket and left for 15 s. They were placed in 1.5 ml of RTF (Reduced Transport Fluid) (24). Each sample had 8 paper points that were processed immediately. Samples were then stirred vigorously for 45-60 s, and serial dilutions in RTF were prepared. 100 µl was taken from the sample and placed in an eppendorf tube with 900 µl of RTF. (1st dilution: 1:10). 100 µl was streaked, expanding the sample with a glass rod on a TSVB plate: tryptic soy serum, bacitracin (75 µg/ml), vancomycin (5.0 µg/ml), 10 ml of horse serum (10%) to isolate Aggregatibacter actinomycetemcomitans. The plates were placed in a jar with a CO2-enriched atmosphere, and incubated for 7 days at 37ºC. 100 µl was taken from the RTF original sample and placed in 900 µl of RTF. 100 µl was taken from this tube and placed in another tube with 900 µl (2nd dilution: 1:100).100 µl was taken and streaked in base agar medium with blood with menadione and hemin, and incubated for 14 days at 37ºC in absolute anaerobiosis. From the original sample in RTF, 500 µl was taken, stored at -30ºC and then used in the multiplex-PCR.
The following strains were used as positive controls: Aggregatibacter actinomycetemcomitans (ATCC 29522), Porphyromonas gingivalis (BAA-308), Prevotella intermedia (ATCC 25611), Tannerella forsythia (ATCC 43037) and Fusobacterium nucleatum (ATCC 25586), and as negative control the Escherichia coli strain (ATCC 47076).
Identification and enumeration of colonies
Presumptive colonies of A. actinomycetemcomitans were identified given the presence of a star-like structure inside the colonies (stereo magnifier), Gram stain technique, catalase test (+) and MUG negative (4-Methylumbelliferyl-β-D-galactoside to study lactose fermentation). We calculated the total number of colony-forming units per milliliter in each sample (CFU/ml) from the streaked culture media. The relative recovery percentage was calculated from the total CFU times the dilution factor. The enumeration of colony-forming units of pigmented anaerobic bacteria was performed in each sample. In general, no species of the pigmented colonies were identified, and all pigmented CFU were counted by dilution factor.
Control strains
Metagenomic DNA extraction from oral microbiota
Samples were defrosted and homogenized for 30 s with a vortex mixer. They were then transferred to another eppendorf tube and centrifuged at 13.500 rpm for 3 min. The supernatant was discarded and the pellet was resuspended again by pipetting in 500 µl of RTF buffer for 5 min at 100ºC. They were then placed in ice for 5 min and then centrifuged at 13.500 rpm for 5 min at 4ºC. Finally the pellet was discarded and the supernatant with the microbial metagenome was stored at -30ºC until multiplex-PCR was performed.
Extraction of bacterial genomes and selection of oligonucleotides
The extraction of the genome of each control strain was done using the ZymoBead™ Genomic DNA Kit (as described by manufacturers). DNA concentration (ng/µl) in each strain was quantified in NanoDrop 2000.
In the selection of species-specific oligonucleotides we consulted bibliographical sources with information on species-specific oligonucleotides of the 16S ribosomal RNA in each positive control strain of this study. Bases were added to the sequences of some oligonucleotides to optimize the conditions of the multiplex-PCR (Table 1 - Figs. 1, 2, 3).
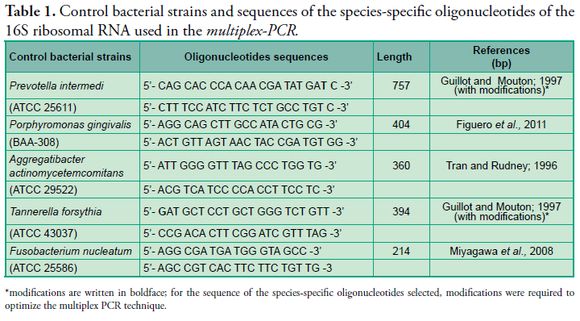
*modifications are written in boldface; for the sequence of the species-specific oligonucleotides selected, modifications were required to optimize the multiplex PCR technique.
ATCC = American Type Culture Collection.
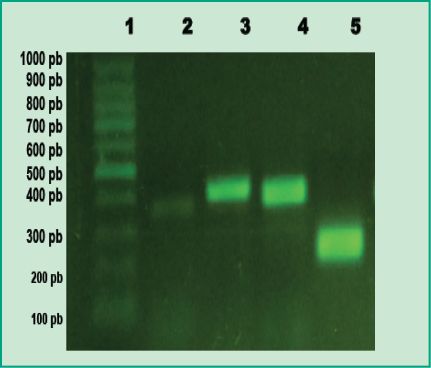
Figure 1: PCR products in agarose gel: Lane 1, 100 bp molecular weight marker
(DMF-50, sbs); Lane 2, positive control of A. actinomycetemcomitans; Lane 3, positive control of P. gingivalis; Lane 4, positive control of T. forsythia; Lane 5, positive control of F. nucleatum
(Agarose gel 1%, GoodView 5%).
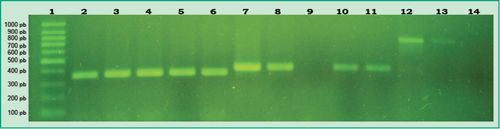
Figure 2: PCR products in agarose gel: Lane 1, 100 bp molecular weight marker (DMF-50, sbs); Lane 2, positive control of A. actinomycetemcomitans; Lanes 3-6, samples from different patients that tested positive for A. actinomycetemcomitans; Lane 7, positive control of P. gingivalis; Lane 8, patient sample that tested positive for P. gingivalis; Lane 9, patient sample that tested negative for P. gingivalis; Lanes 10-11, samples from different patients that tested positive for P. gingivalis; Lane 12, positive control of P. intermedia; Lane 13, patient sample that tested positive for P. intermedia; Lane 14, patient sample that tested negative for P. intermediate
(Agarose gel 1%, GoodView 5%).
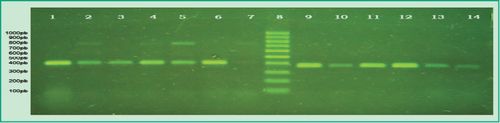
Figure 3. Multiplex-PCR products in agarose gel: Lane 1, positive control of P.gingivalis; Lane 2, patient sample that tested positive for P.gingivalis and P. intermedia; Lane 3, patient sample that tested positive for P.gingivalis and negative for P. intermedia; Lanes 4-5, patient samples that tested positive for P. gingivalis and P. intermedia; Lane 6, patient sample that tested positive for P.gingivalis and negative for P. intermedia; Lane 7, patient sample that tested positive for P.gingivalis and P. intermedia; Lane 8, 100 bp molecular weight marker (DMF-50, sbs); Lane 9, positive control of T. forsythia; Lanes 10-14, patient samples that tested positive for T. forsythia
(Agarose gel 1%, GoodView 5%).
Determination of detection limit
The lowest detection limit was defined as the smallest number of microorganisms in a sample that could be detected by the multiplex-PCR. This was done through serial dilutions (0 to 1.75x106 cells/ml) of a mixture of pure cultures of all the strains described above.
Multiplex-PCR
Programming conditions were optimized according to the parameters of each sequence of oligonucleotides designed in this study (nt, G + C%, Tm). Each reaction mixture (25 µl) contains 1.0X PCR buffer (10 mM Tris-HCl, 50 mM KCl, pH 8.3), 1.5 µl of 2.5 mM dNTPs, 1 mM MgCl2, 10 pmol of each oligonucleotide (Table 1, Figs. 1, 2, 3) and 1.2 U of Taq DNA polymerase and 5 ng of the metagenoma extracted from each sample and 2 ng of the genome of the control strains (NanoDrop 2000).
In the detection by multiplex-PCR of the P. gingivalis and P. intermedia species, the following conditions were applied: 1 cycle of denaturation at 95ºC for 3 min, 35 cycles of 94ºC for 1 min, 55ºC for 1 min, 72ºC for 2 min, and 1 cycle of final extension of 72ºC for 5 min.
The following conditions were applied in the detection by multiplex-PCR of the species A. actinomycetemcomitans, T. forsythia and F. nucleatum: 1 cycle of denaturation at 95ºC for 2 min, 35 cycles of 94ºC for 30 s, 55ºC for 1 min, 72ºC for 2 min, and 1 cycle of final extension of 72ºC for 5 min.
The results of amplifications (bands) were viewed in 1.2% agarose gel, 0.5 X TBE (Tris-borate-EDTA) buffer, 5% of GoodView, 1% of Cyan/Orange loading buffer; 2 µl of the PCR product for 2 h at 80 V. The results were recorded using a photodocumentation device (PHOTO/Phoresis; Fotodyne, UV).
Statistical analysis
The results of the multiplex-PCR were obtained using absolute frequencies in percentages, and the clinical conditions were compared by means of the Student’s-T Test with a significance level of < 0.05.
Results
A total of 51 samples collected from patients with chronic periodontitis were processed with the conventional method of microbiological cultures. The results of this study showed that 23% of patients had A. actinomycetemcomitans in values of 3 x 105 UFC/ml, and 10% in a range of 2-7 x 104 UFC/ml. It was determined that 100% of the samples had black-pigmented anaerobic bacteria (BPAB) in the following percentages: 14% had counts lower than 105 UFC/ml, 74% between 105 - 106 UFC/ml, and finally 12% had counts higher than 106 UFC/ml of the sample. These results showed that there was confirmed presence of BPAB in the periodontal pockets of all the patients. If we remember that in the group of the BPAB quantified in the cultures we can probably find some genera of Porphyromonas, Prevotella and Fusobacterium, then, there may be adhesion, aggregation, and coaggregation among these genera that form the gingival microbiota. For example, it has been shown that F. nucleatum can participate in coaggregation through the interaction of certain lipopolysaccharides which recognize and join the surface O-galactosides of A.actinomycetemcomitans, P. gingivalis and P. intermedia (25, 26). This is of great importance given the possible “bridge” role played by F. nucleatum in the colonization of the subgingival area. F. nucleatum can survive high concentrations of oxygen and is one of the most important bacterial species that works as a bridge between aerobes and obligate anaerobes, actively participating in the link and establishment of the microbial consortium (27).
The results obtained by simultaneous detection of six oral bacterial pathogenic species from 51 patients by multiplex-PCR showed that 100% of the samples tested positive for F. nucleatum; this shows that it is the most prevalent species in a Uruguayan population with chronic periodontitis (Fig. 4).
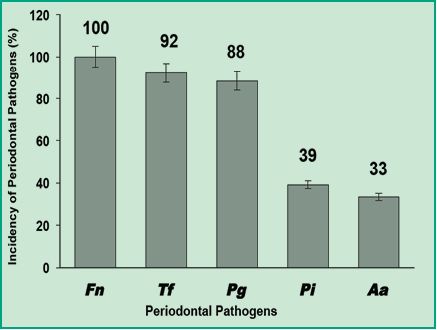
Figure 4. Graph with the incidence percentages of periodontal pathogens in Uruguayan patients with chronic periodontitis.
These results indicate that the most prevalent species is F. nucleatum, the same as in the studies conducted in Chile (11). This information is very relevant as it has been shown that F. nucleatum can colonize the colon and become a prominent biological factor in the development of colorectal carcinoma. This type of cancer causes approximately 610.000 deaths annually around the world and is connected with infection by F. nucleatum, which occurs mainly orally (6). Additionally, T. forsythia and P. gingivalis pathogens were also highly prevalent with 92% and 88% respectively. This shows that these are high-frequency pathogens in a Uruguayan population with chronic periodontitis. When comparing this with the studies conducted in Colombia and Chile (1, 11) we can state that P. gingivalis also has a high prevalence in Uruguay. The high prevalence of both species is important because these are endogenous pathogens, that is to say that they can invade the epithelial tissue and allow entry to other opportunistic microorganisms such as Herpes virus, Candidas spp., among others. On the other hand, P. intermedia and A. actinomycetemcomitans were detected less frequently: 49% and 33%, respectively (Fig. 4). Depending on prevalence percentages, Gajardo and colleagues used the following criterion: the higher the percentage of detection of a pathogenic species, the higher the link between the pathogen and the development of chronic periodontitis. According to this criterion, in this study we consider F. nucleatum, T. forsythia and P. gingivalis as critical pathogens in connection with chronic periodontitis, and P. intermedia and A. actinomycetemcomitans as bacteria associated with chronic periodontitis with lower prevalence.
We must highlight the strong evidence regarding the potential association between periodontal diseases and the development of cardiovascular complications in both men and women (28-30). Therefore it is important to implement fast detection and treatment of patients, thus avoiding the action of these invasive pathogens that can favor the adhesion of leukocytes to vascular epithelium, which might have a pro-coagulant effect among others, as has been reported in the bibliography (3).
When comparing our results with the results achieved Chile (1, 11), Spain, Colombia (31) and Brazil (14), we can observe that the most prevalent species vary in each country. This might be due to various factors such as the existence of geographical, racial, etiological, genetic, environmental, habit-related and oral health behavior differences.
The significant differences detected between men and women might be due to the different factors and mechanisms involved in the events of colonization of the periodontal pockets in both sexes. Besides, it is highly likely for the establishment and multiplication of a pathogenic species to be different from that of another pathogenic species; or a bacterial species can be favored by the environmental conditions of a host but not of other hosts. Klinger and colleague established that nutrients available to the bacterial consortium inside periodontal pockets are very different in men than in women, which is mainly determined by hormonal levels (32). Ethnic origin can probably influence the selection of bacterial species in the gingival microbiota. Besides, reports have shown that some have a predilection for a specific ethnic group, for example, P. gingivalis is found mainly in African-American patients with periodontitis; while F. nucleatum seems to be more frequent in Caucasian patients.
Figure 5 shows the values of simultaneous and combined detection by order of incidents of the five periodontal pathogens present in both female and male patients. Interestingly, men had higher simultaneous detection values than women. Besides, among men, the co-detection of F. nucleatum and T. forsythia reached 100%, while among women it was significantly lower (83%). The same trend was observed in the simultaneous detection of the three pathogens F. nucleatum, T. forsythia and P. gingivalis: 93% among men and 70% among women. As simultaneous detection increases (F. nucleatum, T. forsythia, P. gingivalis and P. intermedia), the percentages of detection diminish; nevertheless, they remain significantly higher among men (39%) than among women (26%). Finally, the simultaneous detection of all the periodontal pathogens of this study (F. nucleatum, T. forsythia, P. gingivalis, P. intermedia and A. actinomycetemcomitans) showed values of 18% among men and 13% among women (Fig. 5). These results suggest that subgingival microbiota greatly differs between men and women of a Uruguayan population with chronic periodontitis. In men there are multigeneric bacterial coaggregations, perhaps especially favored by the hormonal microenvironment, different from that of women.
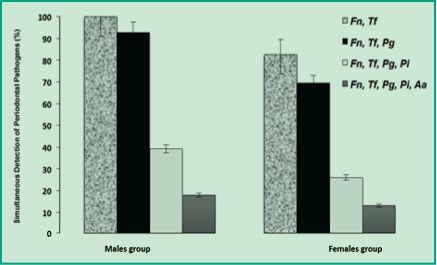
Figure 5. Percentages of multiple detection of periodontal pathogens in men and women
Conclusions
The samples analyzed using the conventional microbiological technique showed a 33% presence of A. actinomycetemcomitans and a 100% presence of black-pigmented anaerobic bacteria. The analysis of the species-specific sequences of 16S ribosomal RNA with mutiplex-PCR allowed for the fast detection, in each species, of five periodontal pathogens in samples taken from 51 patients of both sexes. Among the bacterial species identified, F. nucleatum was the only periodontal pathogen detected in all male and female patients. The most prevalent pathogenic species in a Uruguayan population with chronic periodontitis were F. nucleatum, T. forsythia and P. gingivalis.
Acknowledgments
We would like to thank the technical support of the School of Graduates of the School of Dentistry, Universidad de la República, as well as Paul Gaytán, Jorge Yáñez and Eugenio López for the synthesis of oligonucleotides, IBT- UNAM (Mexico).
1. Gajardo M, Silva N, Gómez L, León R, Parra B, Contreras A, Gamonal J. Prevalence of Periodontopathic Bacteria in Aggressive Periodontitis Patients in a Chilean Population.J. Periodontol. 2005;76: 289-294.
2. Bertl K, Zatorska B, Leonhard M, Matejka M, Schneider-Stickler B. Anaerobic and microaerophilic pathogens in the biofilm formation on voice prostheses: A pilotstudy. Laryngoscope. 2011; 6. 10.1002/lary.23193. [Epub ahead of print].
3. Figuero E., Sánchez M., Cuesta S., Tejerina J., Del Castro J., Gutiérrez J., Herrera D., Sanz M. Detection of Periodontal Bacteria in Atheromatous Plaque by Nested Polymerase Chain Reaction. J. Periodontol.2011; 82(10): 1469-1477.
4. Byakodi R, Krishnappa R, Keluskar V, Bagewadi A, Shetti A. The microbial flora associated with oral carcinomas. Quintessence Int.2011;42(9): 118-123.
5. Cao H, Qi Z, Jiang H, Zhao J, LiuTang. Detection of Porphyromonas endodontalis, Porphyromonas gingivalis and Prevotella intermedia in primary endodontic infections in a Chinese population. Int. Endod. J. 2012 Mar 19. doi: 10.1111/j.1365-2591.2012.02035.x. [Epubahead of print].
6. Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res.2012; 22(2): 299-306.
7. Albandar JM. Periodontal diseases in North America. Periodontol 2000.2002; 29:31-69.
8. Bascones-Martinez A F. Las enfermedades periodontales como infecciones bacterianas. Av Periodon Implantol.2005;17(3):147-56. (Albandar; Kleonatas et al.; Ghizoni et al.).
9. Kleontas A, Asteriou C, Efstathiou A, Konstantinou E, Tsapas C, Barbetakis N. Actinomyces israelii: a rare cause ofthoracicempyema. Tuberk Toraks.2011; 59(4): 399-401.
10. Ghizoni JS, Taveira LA, Garlet GP, Ghizoni MF, Pereira JR, Dionísio TJ, Brozoski DT, Santos CF, Santana AC. Increased levels of Porphyromonas gingivalis are associated with ischemic and hemorrhagic cerebrovascular disease in humans: an in vivo study. J. Appl. Oral Sci.2012; 20(1): 104-112.
11. Mujica TC., Castillo-Ruiz M., Daille LK., Fuentevilla A., Bittner M. Co-detection of Periodontal Pathogens in Chilean Patients with Chronic Periodontitis. Rev. Clin. Periodoncia Implantol. Rehabil. Oral.2010;3(3): 118-122.
12. Fayad I., Lafaurie G., Contreras A., Castillo D., Barón M. Microflora subgingival en periodontitis crónica y agresiva en Bogotá, Colombia: un acercamiento epidemiológico. Biomédica., 2007;27(1);16-20.
13. Medina A., Montoya A., Zuluaga G. Perfil microbiológico subgingival de pacientes con periodontitis crónica en una población de Colombia. Avances en Periodoncia.2012;24(1): 47-53.
14. Taba M Jr., Souza SL., Mariguela VC. Periodontal disease: a genetic perspective. Braz Oral Res.2012; 1:32-38.
15. Papone V, Batlle A. Porphyromonas gingivalis. Su relación con Periodontitis Rápidamente Progresiva. Odontología de Postgrado.1997;3(4): 51-55.
16. Papone V, Morteo G. Un patógeno periodontal virulento Actinobacillus actinomycetemcomitans. Actas odontológicas .2005;2(1): 43-47.
17. Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products.Chemistry&biology.1998;5(10), R245-R249.
18. Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, Loiacono KA, Lynch BA, MacNeil IA, Minor C, Tiong CL, Gilman M, Osburne MS, Clardy J, Handelsman J, Goodman RM. Cloning the Soil Metagenome: a Strategy for Accessing the Genetic and Functional Diversity of Uncultured Microorganisms. Appl.Environmental Microbiology.2000;66(6): 2541-2547.
19. Mullis KB, Ferré F, Gibbs R. The Polymerase chain reaction. Optimization of Multiplex PCRs. Ed. Birkhäuser.1994; Boston. 38-54.
20. Tran SD, Rudney JD. Multiplex PCR Using conserved and species-specific 16S rRNA gene primers for simultaneous detection of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. J. Clin. Microb.1996; 34 (11): 2674–2678.
21. Tran SD, Rudney JD. Improved Multiplex PCR using conserved and species-specific 16S rRNA gene primers for simultaneous detection of Actinobacillus actinomycetemconitans, Bacteroides forsythus and Porphyromonas gingivalis. J. Clin. Microb. 1999;35(11):3504-3508.
22. Willis SG., Smith KS., DUNN VL., Gapter LA., Riviere KH., Riviere GR. Identification of Seven Treponema Species in Health- and Disease-Associated Dental Plaque by Nested PCR. American Society Microbiology. 1999;37(3): 867-869.
23. Armitage, G.Development of a classification system for periodontal diseases and conditions. Annals of Peridontology .1999;4, 1-6.
24. Syed SA., Loesche WJ. Survival of Human Dental Plaque Flora in Various Transport Media. App. Microbiology.,1972; 24(4): 638-644.
25. Kolenbrander PE. Multispecies communities: interspecies interactions influence growth on saliva as sole nutritional source. Int J Oral Sci.2011; 3: 49-54.
26. Rosen G, Nisimov I, Helcer M, Sela M. Actinobacillus actinomycetemcomitans Serotype b Lipopolysaccharide Mediates Coaggregation with Fusobacterium nucleatum. Inf Immunity.2003 71(6): 3652–3656.
27. Huang R., Li M., Gregory R. Bacterial interactions in dental biofilm. Virulence.2011; 2(5): 435-444.
28. Bahekar AA, Singh S, Saha S, Molnar J, Arora R. The prevalence and incidence of coronary heart disease is significantly increased in periodontitis: A meta-analysis. J Am Heart.2007; 154: 830-837.
29. Humphrey LL, Fu R, Buckley DI, Freeman M, Helfand M. Periodontal disease and coronary heart disease incidence: A systematic review and meta-analysis. J Gen Intern Med.2008; 23: 2079-2086.
30. Blaizot A, Vergnes JN, Nuwwareh S, Amar J, Sixou M. Periodontal diseases and cardiovascular events: Meta- analysis of observational studies. J Int Dent; 2009;59: 197-209.
31. Herrera D, Contreras A, Gamonal J, Oteo A, Jaramillo A, Silva N, Sanz M, Botero J, León R. Subgingival microbial profiles in chronic periodontitis patients from Chile, Colombia and Spain. J. Clin. Periodontol.2008; 35: 106-113.
32. Klinger G., Glänzer S., Sigusch B., Klinger G., Römer WP. Influence of sexual steroids on cell functions of PMNL in the gingival sulcus. Pharmazie. 2000; 55(9): 678-680.










 text in
text in