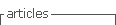Services on Demand
Journal
Article
Related links
Share
CLEI Electronic Journal
On-line version ISSN 0717-5000
CLEIej vol.18 no.2 Montevideo Aug. 2015
A middleware-based platform for the integration of bioinformatic services
Guzmán Llambías and Raúl Ruggia
Universidad de la República, Facultad de Ingeniería,
Montevideo, Uruguay, 11300
Abstract
Performing Bioinformatic’s experiments involve an intensive access to distributed services and information resources through Internet. Although existing tools facilitate the implementation of workflow-oriented applications, they lack of capabilities to integrate services beyond low-scale applications, particularly integrating services with heterogeneous interaction patterns and in a larger scale. This is particularly required to enable a large-scale distributed processing of biological data generated by massive sequencing technologies. On the other hand, such integration mechanisms are provided by middleware products like Enterprise Service Buses (ESB), which enable to integrate distributed systems following a Service Oriented Architecture. This paper proposes an integration platform, based on enterprise middleware, to integrate Bioinformatics services. It presents a multi-level reference architecture and focuses on ESB-based mechanisms to provide asynchronous communications, event-based interactions and data transformation capabilities. The paper presents a formal specification of the platform using the Event-B model.
Abstract in Spanish
La ejecución de los experimentos bioinformáticos involucra el uso intensivo de servicios y recursos de información distribuidos a través de Internet. Las herramientas existentes facilitan su implementación siguiendo una orientación a flujos de trabajo (workflows), pero carecen de capacidades de integración de servicios con patrones de interacción heterogéneos y a gran escala. Esta integración es especialmente necesaria para permitir a un procesamiento distribuido a gran escala de los datos biológicos generados por las tecnologías de secuenciación masiva. Por otro lado, estos mecanismos de integración ya son proporcionados por productos de middleware como los Enterprise Service Bus (ESB), que permiten integrar sistemas distribuidos siguiendo el paradigma promovido por las Arquitectura Orientada a Servicios (SOA). Este trabajo propone una plataforma de integración basada en middleware empresarial para integrar servicios bioinformáticos. En él se presenta una arquitectura de referencia con varios niveles de abstracción y se describen con mayor detalle los mecanismos basados en ESB para proporcionar comunicaciones asíncronas, interacciones basadas en eventos y capacidades de transformación de datos. En el artículo presenta una especificación formal de la plataforma utilizando el modelo de eventos-B.
Keywords: Middleware, Bioinformatics, Integration Platforms, Platform as a Service (PaaS).
Keywords in Spanish: Middleware, Bioinformática, Plataformas de integración, Plataforma como Servicio (PaaS).
Submitted: 2014-11-11. Revised: 2015-04-07. Accepted: 2015-07-12.
1 Introduction
Bioinformatics is a multidisciplinary area involving Biology and Computer science, whose purpose is to get a more clear vision of the organisms’ biology through IT-based tools. The ordered and combined application of these constitutes the so-called in-silico bioinformatic experiments.
An in-silico experiment corresponds to a procedure accessing information repositories and applying local and remote analytical tools to show certain hypotheses, derive results, and search patterns [1]. In turn, Bioinformatics repositories may consist of biological databases like EBML [2] or Swiss-Prot [3], while analytical tools may consist of the implementation of complex algorithms like BLAST [4] and ClustalW [5].
In-silico experiments are nowadays implemented using Workflow Management System (WMS) tools specially adapted to the biological context. In the last years, scientific workflows have revolutionized the way researchers perform their experiments by enabling them to use powerful computational tools without needing a strong IT background. For instance, these tools allows scientists to access external services using Web Services, perform data format transformations, execute queries on large databases and even request the execution of experiments in the cloud. Among others, Taverna [6] is one of the most popular scientific workflow tools. Developed in the context of the myGrid Project, Taverna provides capabilities to model experiments as dataflows, perform implicit communications, collect provenance information and integrate with third parties (e.g. BioMart).
Although the generalized use of WMS tools is a clear success indicator [7], in particular Taverna, it presents important limitations. In particular, the polling approach followed by most biological service providers and the use of different data formats in services makes the logic implemented by workflows to be more complex. Furthermore, it doesn’t provide asynchronous message-oriented mechanisms and quality of services management [8] [9]. In these scenarios, workflow development remains highly complex.
Furthermore, such functionalities are required to respond with more powerful distributed and collaborative systems to the increasing scale of data volumes to be processed, which are generated by “High Throughput” and Next Generation Sequencing (NGS) technologies for massive sequencing [10].
On the other side, enterprise middleware technologies, which have been evolving during the last years, provide abstractions and solutions to increasingly complex integration functionalities (e.g. asynchronous interactions and interoperable interactions over the internet) related to the construction and integration of distributed applications. Particularly, Enterprise Service Buses (ESB) and cloud-based Internet Service Buses (ISB) are sophisticated middleware technology progressively being leveraged to integrate highly distributed and heterogeneous services. ESBs provide mediation capabilities (e.g. message transformation and intermediate routing) which can be used to address mismatches between applications and services regarding communication protocols, message formats, interaction styles and quality of service (QoS), among others [11] [12].
These elements lead to the goals of improving the collaboration in bioinformatics community by enhancing service-based integration capabilities as well as reducing the complexity in the involved development by using tools with more advanced mechanisms than the ones provided by local Workflows (i.e. enterprise middleware technologies). Such enhanced platforms should enable to combine bioinformatics Web-based services (e.g. NCBI Web Services), experiment-oriented workflow tools (e.g. Taverna) with general purpose mediation features (e.g. asynchronous interactions, rules, events, data transformation) and other value-added capabilities (e.g. service policy compliance). The ultimate goal would be to put into practice a Platform as a Service (PaaS) approach in the bioinformatics area enabling an active participation not only of laboratories and researchers, but also middleware platforms and cloud computing services suppliers and developers.
This paper addresses these issues and proposes a reference integration platform for the bioinformatics domain which, mediating between WMSs (e.g. Taverna, Kepler, Galaxy, etc.) and bioinformatics services, provides mechanisms that facilitate the development of distributed scientific workflows and solve common challenges that arise when performing these tasks. The proposed integration platform leverages mediation features of enterprise middleware, particularly ESBs, addresses identified integration requirements by improving asynchronous interactions, event notification and message transformation capabilities, and provides the means to implement other value-added services such as service policy compliance.
This proposal aims at combining Bioinformatic Web Services (e.g. NCBI), tools to develop scientific workflows (e.g. Taverna) and mechanisms to integrated distributed applications through an integration platform based on advanced enterprise middleware. The goal is to enable the integration of scientists, laboratories and bioinformatics service providers to develop sophisticated bioinformatics experiments.
The proposed Integration Platform is based on different type of middleware technologies (Web Services, Message Queues, ESB) and enables to connect workflow tools (e.g. Taverna) and Bioinformatic services (e.g. NCBI) in order to solve the requirements related to implement in-silico experiments. This Integration Platform takes advantage of mediation functions provided by enterprise middleware tools (particularly ESB). This paper also includes a formal specification of the platform using the Event-B model. This Integration Platform, which is specific to the Bioinformatic domain, is characterized by three dimensions: (i) functionalities required to solve the requirements related to in-silico experiments, (ii) the application scenario given by the type of Bioinformatic Laboratory, and (iii) the type of middleware technology to be used.
This paper is based on the Master Thesis [13]. A more detailed description of the topics may be found in the thesis document.
The rest of the paper is organized as follow. Section 2 presents background on Bioinformatic tools and ESB middleware as well as a description of Related Work. Section 3 describes the proposed Integration Platform identifying requirements as well as application scenarios, and presenting a high-level architecture. Section 4 presents a use case example of the platform. Section 5 presents a formal specification using Event-B model. Section 6 describes an ESB-based implementation of the Platform. Section 7 presents conclusions and future work.
2 Existing Knowledge and Related Work
2.1 Bioinformatic Concepts and Project myGrid
Bioinformatics is a multidisciplinary area involving Biology and Computer Science, which performs the so-called in-silico experiments through a combined application of IT tools and by accessing to local and distributed data. For instance, an experiment researching on the evolutions of relationships between proteins may start acquiring an amino acid sequence from the Swiss-Prot repository and afterwards applying to it the ClustalW algorithm to align sequences and to identify patterns among these last [1] [2] [3] [4] [5] [14].
At the beginning, performing in-silico experiments was based on “copy & paste” of data in Web-forms, afterwards evolving to Perl and Python scripts with screen-scrapping techniques. Later on, the advent of Web Services technologies enabled a programmatic-based access to services provided by “third parties”. This approach was strengthen with tools like Biological WMS (Workflow Management Systems), which enabled biologists to model their experiments as workflows composed by multiple Bioinformatic Web Services [14].
These tools provide biologists with required abstractions to implement in-silico experiments accessing to databases, using technologies such as Web Services and Cloud-based tools [6] without needing a high expertise on IT.
These Bioinformatics techniques and tools have enabled biologists to carry out experiments in an alternative way to the traditional in-vitro one, facilitating the reuse of results obtained worldwide by reusing through data reuse in in-silico experiments.
By 2005 emerged the Next Generation Sequencing (NGS) technologies, which will generate a revolution in sequencing techniques. Among others, NGS-based projects include de novo genome sequencing, transcriptome analysis, and variability analysis. Equipment performing such sequencing, 454 Roche and Illumina among others, generate much larger data volumes, in shorter times and with lower costs than the preceding High-Throughput technologies. For example, the Illumina HiSeq 2000 may produce two hundred millions of “paired-end reads” (200 Gb) in each execution [10].
In this context of very large data volumes, the traditional bioinformatics scenarios based on downloading data and processing it locally are no longer viable given the limited scalability. For instance, executing a BLAST or analysis or even a BWA analysis (high-speed mapping algorithm) [15] would take several days, which is completely unsuitable for scientists. In addition, transferring these data constitutes a challenge by itself and it’s more susceptible to errors, specially due to weaknesses in existing protocols to transfer very large data volumes [16].
As a consequence, this new technology brought notable improvements but also involved new challenges, completely different to the ones identified in the former generation of experiments [17].
In this new context, applying Cloud Computing approaches appears to be promising due to elasticity capabilities to store and process very large data volumes. However, these features are not sufficient to address these issues, and other problems remain open, such as security, data transfer [18], in addition to defining the necessary abstractions enabling an appropriate use of Cloud-based capabilities, as well as to simplify the development of the in-silico experiments [19].
2.1.1 myGrid Project
myGrid [20] is an e-Sciences project started in 2001 aiming to provide workflow-oriented tools using basic middleware mechanisms (i.e. Web Services) that can simplify the development of biological and bioinformatics experiments. Some of the tools developed by this project are:
-
Taverna: a tool that allows scientists to model their experiments as composition biological services and build scientific workflows
-
myExperiment: provides a collaborative environment to scientists, enabling the publication, “curation” and search of experiment workflows.
-
BioCatalogue: an open biological Web Service registry to enable the publication, classification and search of Web Services for the scientific community.
All these components were developed following a SOA paradigm and using Web Services, Workflows, Web 2.0 and Semantic Web technologies. The components follow an open-source approach and may be extended by third-parties. Therefore they provide appropriate means to build the local platform to be connected to distributed and virtual research environments, the so-called e-Labs [21].
The main contribution of Taverna is that allowed scientists with limited IT expertise, be capable to develop their experiments using advanced technologies such as SOAP Web Services, BioMart, SoapLab, R or SADI services.
2.2 Enterprise Middleware and the Enterprise Service Buses
Generally speaking, middleware technologies are general purpose infrastructure services positioned between applications and/or platforms, which allow their interactions systematically using high productivity tools. These technologies have had an impressive evolution in the last years, emerging different types of products that enable to achieve multi-platform system integration at different scales. The most remarkable are the Web Services [22], Message Queues and Enterprise Service Bus (ESB) [23], which will be used in this work.
The most advanced technologies, especially ESBs, possess mediation capacities (e.g. message routing and transformation), which may be defined as a “layer of intelligent middleware services that enable to connect applications and data” by solving a number of issues related to such integration (e.g. data transformation and synthesis) [24]. In addition, ESB enable to solve, in a unique tool, both synchronic and asynchronous application integration through robust and scalable mechanisms.
An ESB is an environment belonging to the platform middleware systems category, which provides sophisticated interconnectivity between services and enables to overcome issues related with reliability, scalability and communication. Service interaction using an ESB is based on a combination of the patterns: Asynchronous Queuing, Event-Driven Messaging, Intermediate Routing, Policy Centralization, Reliable Messaging, Rules Centralization and Transformation. Additionally, there exists a set of interactions styles, which defines the way each actor may behave using an ESB [25].
In this section we will briefly review some of the most useful patterns and styles for this paper.
2.2.1 ESB Design Patterns
Application interactions in ESB may be conceptualized through certain design patterns [25]: Asynchronous Queuing, Event-Driven Messaging, Intermediate Routing, Policy Centralization, Reliable Messaging, Rules Centralization and Transformation. This work mainly applies the following ones:
-
Intermediate routing patterns dynamically determine the message path according to different factors. More concretely, a content-based router defines the message path based on its content and a recipient list routes the message to a list of dynamically specified recipients. Other basic routing patterns include context-based router, load balancing router, aggregator, message filter, static router, dynamic router, splitter, resequencer and discovery.
-
Transformation patterns deal with the runtime transformation of messages. In [25] the authors identify three types of transformations: data model transformations, data format transformations and protocol bridging. Additionally, in [26] various transformation patterns are identified including envelop wrapper, content enrichment, content filter, claim check, normalize and canonical data model.
-
The Asynchronous Queuing pattern deals with the interactions of client and services where synchronous communication can inhibit performance and compromise reliability.
-
Event driven messaging determines an event driven based exchange of messages between systems where the receiver receives messages according to the events it is subscribed to.
2.2.2 Interaction Styles
In addition, the “interaction style” patterns define the way actors behave and interact in terms of synchronous/asynchronous interfaces, level of assurance of delivery, handling of timeouts, late responses, and error handling in an ESB-like middleware [27].
In this work, we will focus on two interactions styles: 1) Synchronous update request with acknowledgement using callback operations, and 2) One-way with status poll for completion.
A one-way request with status poll for completion consists of an initial synchronous response-less request followed by periodic synchronous requests from the requester to know the status and result of the operation. In turn, a synchronous update request with acknowledgement using callback operation enables the requester to be sure that the request was received and is being processed without being blocked. The response is received through another synchronous communication from the invoked service (the callback).
2.3 Domain Specific Middleware Platforms
Domain-specific middleware platforms provide implementation building blocks and services adapted for the requirements of a specific domain; they provide applications with reusable abstractions of recurring patterns and algorithms [28]. Therefore they constitute a relevant knowledge background in the present work, particularly the ones on two domains: Health and geographic-based services in e-Government.
The Open eHealth Integration Platform (IPF) [29] is a platform middleware for the e-health domain. It has the goal of providing the required integration mechanisms (e.g. HL7 message processing, support for the IHE profiles) to enable the integration of applications in that domain.
In [30] the authors propose an ESB-based integration platform in the context of enterprise geographic information systems. The authors propose a series of mediation mechanisms (e.g. SOAP WMS-Wrapper, WMS Enricher) to facilitate the integration of geographic Web Services and enterprise applications.
To the best of our knowledge there are not proposal of a domain-specific integration platforms in the bioinformatics domain.
2.4 Event-B
Event-B, the evolution of B method, is a formal method including a language and a tool to support the verification of properties on the specifications [31]. In Event-B, systems are formally specified as Abstracts Virtual Machines (AVM o MACHINE), which have a structure based on the VARIABLES constructor and a dynamic behavior based on events (EVENT constructor). In addition, conditions and event execution and values of variables may be specified through the INVARIANTS constructor.
Among a large number of applications, Event-B has been used to formalize distributed transactions management systems [32] as well as middleware software [33].
2.4 Related Work
This section presents related work of Bioinformatic applications aiming at providing different integration mechanisms for in-silico experiments.
The need for asynchronous interactions in the context of scientific workflows is not new and there are different proposals which use WS-Addressing in order to provide solutions for this requirement.
The work in [34] proposes a middleware for asynchronous interactions based on WS-Addressing, which is integrated to Taverna by means of a new processor called “Generic Web Service Processor”. In turn, our proposal does not depend on this feature.
The proposal in [35] follows a similar approach by proposing extensions to WS-BPEL for allowing the use of WS-Addressing. Both works have the limitation of assuming that biological Web Services has support for WS-Addressing which, in practice, is not that frequent. Indeed, the main providers (e.g. EBI, EMBL, DDBJ, GenBank) do not yet provide Web Services with WS-Addressing support and adopt the one-way request with status poll for completion approach for asynchronous interactions. On the contrary, our approach does not require that services have WS-Addressing support: it leverages existing services and adapts them so they can be invoked using WS-Addressing (e.g. by solutions like [34] and [35]).
The use of notification mechanisms and events has also been addressed in the bioinformatics domain. In [35] Apache ODE, a WS-BPEL engine, is extended to send notifications according to events of interest in an e-science context. Compared to our approach, this solution has the limitation to be coupled to the WS-BPEL engine, not allowing it to be used in wider scenarios (e.g. the advanced types of laboratories of Section 3.3).
In [36] the authors propose a Web Service based message broker, called WS-Messenger, with the goal of sending notifications of events (logging, monitoring, etc.) among applications, platforms and heterogeneous grid environments. Similar to our proposal, WS-Messenger is based on standards (e.g. WS-Eventing and WS-Notification) and provides some mediation capabilities at the transport level. The main difference with our notification mechanism is the scale of the solution: while WS-Messenger is a lightweight middleware suitable to be integrated in other platforms, our mechanism is built on top of an ESB infrastructure. Also, our mechanism is part of a wider integration platform for the bioinformatics domain which facilitates its integration with the rest of the services and infrastructure deployed in the platform for this domain.
In [37] a WS-Eventing middleware with the goal of improving the interoperability between workflows tools is presented. The solution, called PS-SWIF, provides the same notification functionality that our notification mechanism. However, PS-SWIF only allows synchronous subscriptions while our mechanism also support asynchronous subscriptions.
In [38] the authors propose the use of a notification bus to which all components are connected as well as subscribed to one or more topics. The main difference with our notification mechanism is that ours is based on a service oriented design and it leverages an ESB infrastructure, facilitating in this way the interoperability among distributed and heterogeneous services.
Format transformation has also been addressed in the literature. In [39] the concept of shim service is introduced and the problem of excessive use of shim services is presented and addressed. In particular, the authors propose the use of ontologies to describe Web Services and, in this way, automatically identify which shim services (from an existing library) are required, so that they can be included in the workflow. On the contrary, our work propose reducing, as much as possible, the use of shim services by encapsulating existing services through services which use a predefined data model.
In [40] the concept of Virtual Data Assembly Lines (VDAL) is presented as a way of hiding the use of shim services to scientists. The authors propose encapsulating existing domain services in VDAL, which include the required configuration to solve syntactic and semantic mismatches. Although our proposal is similar, VDALs are locally defined in workflows by the workbench, while our proposal encapsulates services in new services which are global to all the organization.
A set of good practices based on semantic technologies and Web Services to reduce format transformation issues are proposed in [41]. While these practices take advantage of the potential use of RDF, they are still limited as very few Bioinformatic services include RDF descriptions.
Concerning the application of “policies of use” for services, EntrezAjax [42] is one of the few systems that enable to consume Bioinformatic services applying NCBI’s policies, but doesn’t include similar mechanisms for other providers (e.g. EBI, DDBJ).
Additionally, all the previous described work addresses specific problems when developing scientific workflows. In contrast, our approach has the goal of providing a comprehensive domain-specific integration platform for bioinformatics services, which can be adapted or extended (using middleware of different scale) according to the particular requirements and capabilities of the specific laboratories.
3 A Middleware-based Platform for the Integration of Bioinformatics Services
3.1 General Description
An environment for in-silico bioinformatics experiments includes, on one side, local software tools (i.e. Taverna) that enable researchers to develop their experiments through the composition of remote bioinformatics resources. On the other side, there are bioinformatics resources distributed all over the Internet by Bioinformatics Web Services providers (i.e. NCBI, EBI, DDBJ) that can be accessed through SOAP or REST Web Services APIs.
Although bioinformatics tools like Taverna provide very powerful user oriented development features, their lack on mediation mechanisms (e.g. asynchronous interactions, event management, declarative data transformations, etc.) and platform management capabilities (e.g. quality of services, service policy, etc.) limits their integration capabilities and delegate advanced computing tasks to the scientists.
The proposed bioinformatics Integration Platform presented in Fig. 1, allows developing in-silico experiments through traditional synchronous service composition, and also provides new features to cope with advanced requirements that involve asynchronous interactions based on request-response patterns and data transformation.
Local bioinformatics tools may send messages to the Platform, which will be routed using content-based routing components and specific adapters to forward messages to remote bioinformatics services developed with heterogeneous technologies (i.e. SOAP, R, etc.). Response messages will be sent back using callbacks invocations or notifications mechanisms. In this way, the platform hides implementation and physical distribution aspects of the bioinformatics services, enabling scientists to focus on business aspects of applications and not in infrastructure or technical details.
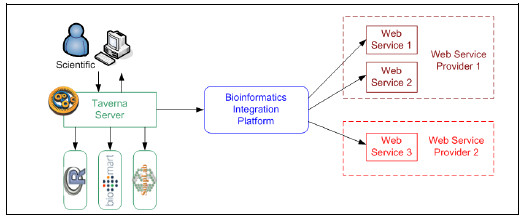
Figure 1: Integration Platform for bioinformatics services
The specification of the integration platform consists of several refinement levels, shown in Fig. 2. The highest level of specification, is independent of technical approaches or laboratory application contexts and specifies the integration platform in an abstract way, integrating local bioinformatics tools (i.e. Taverna or Kepler) with remote bioinformatics services. The second level of specification refines the latter one, introducing refinement dimensions to define more specific Platforms, taking into account middleware technologies, integration requirements (features) and usage contexts scenarios. Finally, the third level of refinement introduces middleware specific products and development details to complete the specification and allow the definition of a concrete instance of the Platform.
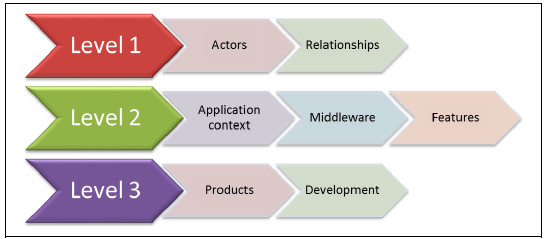
Figure 2: Specification of the Platform in levels of refinements and dimensions for each level
To sum up, refining the top-level specification through refinement dimensions enables the specification of a concrete Platform instance that includes specific technical characteristics. For example, an instantiation of the Platform could cover all functional requirements using Web Services, while another instance may cover the same functionalities but using another type middleware, for example an ESB.
The following subsections will first focus on the new features to solve advanced integration requirements and secondly, it will describe the usage contexts scenarios. Finally, the refinement process is explained in detail using the dimensions explained. A formal specification using Event-B is later on described in section 5.
To illustrate the use of local bioinformatics tools, we use Taverna Workbench as an example, but the Platform is independent of the selected workflow tool and it can be replaced by any other with similar features, like Kepler, Triana or Pegasus.
3.2 Integration Requirements
After analyzing the features required by an environment for bioinformatics in-silico experiments and the actual support of this features by bioinformatics workflows tools [8] [13], we conclude that Taverna does not provide adequate support for a set of relevant features in advanced integration scenarios, namely: asynchronous interactions, event notification, message transformation, service composition and service policy compliance.
Another conclusion of this analysis is that the introduction of middleware technologies allows implementing these features in a relatively straightforward manner.
In this section, we present a resume of the results and a brief description of how these features are provided by the Integration Platform.
3.2.1 Asynchronous Interactions
One of the objectives of this work is to improve asynchronous interactions between bioinformatics services. Nowadays, asynchronous interactions are based primarily on a polling model where “callers” invoke services to submit jobs and later on, “callers” periodically invoke the service for the results (Fig. 3). Besides this model is effective to provide asynchronous interactions, is not the most efficient, as it consumes unnecessary resources for both, processing and communication.
To improve this behavior we propose two solutions: 1) a model based on Web Services using callback invocations or 2) the usage of message queues subscription (topics) to receive the responses of the "invoked" services when they are available. Both solutions are more efficient regarding processing and communication without sacrificing the effectiveness of the interaction (Fig. 4).
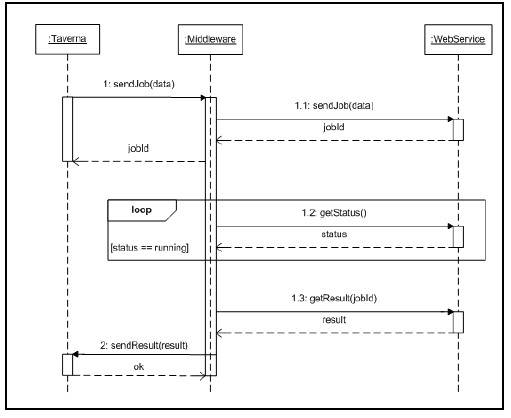
Figure 3: Asynchronous interactions based on a polling to receive responses.
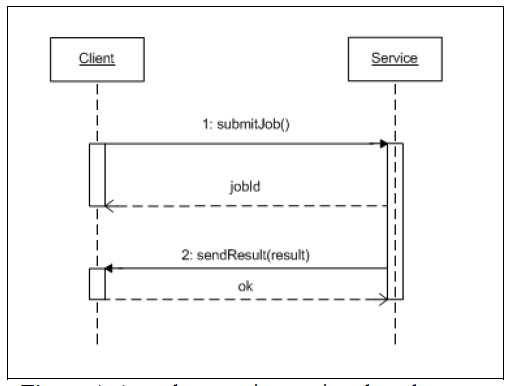
Figure 4: Asynchronous interactions based on callback invocations to receive responses.
The proposed solution, when instantiating the Platform with an ESB middleware (Fig. 5), consists on applying the following design patterns: Asynchronous Queuing (ESB message queues), Intermediate Routing (Content Based Router component of the ESB) and Protocol Bridging (ESB Endpoints and Connectors) [25].
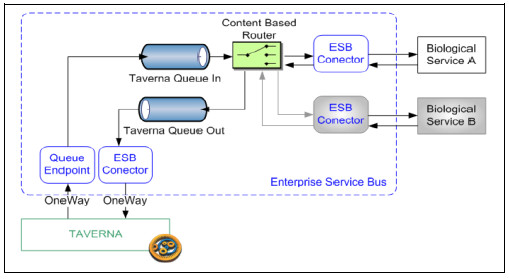
Figure 5: Asynchronous interactions in the ESB
This solution includes two message queues in the ESB: one used to receive messages from Taverna (TavernaQueueIn) and another one, used to send response messages from the ESB to Taverna (TavernaQueueOut). The unique subscriber of TavernaQueueIn queue is a content based router (CBR), which routes messages to its destination according to the message content. For this being done, all messages must specify a target service, which must be registered with in the Platform’s Service Registry. The communication between the ESB and the bioinformatics service is done using ESB Connectors.
After processing the message, the service sends the reply message to the TavernaQueueOut queue, whose sole subscriptor (an ESB connector) will re-sent the message to Taverna. Since there may be multiple instances of Taverna waiting for response messages, a replyTo attribute must be defined in the request messages to specify the destination of the response queue.
3.2.2 Events Notification
In a bioinformatics in-silico experiments environment, several important events may occur and must be detected and used by other bioinformatics systems. An experiment that finishes, messages received from external services, business timeout events (not communication timeouts) and the update notifications of biological databases are some examples.
The proposed ESB-based Integration Platform provides new features to the bioinformatics environment to enable event notifications from Taverna to external services and vice versa. The middleware allows the definition of event topics where services can publish event messages they want to notify.
On the other side, interested services may subscribe to event topics to start receiving event messages published on the topic. These services are called Subscriptors and may choose between two subscription types: synchronous subscription and asynchronous subscription. On asynchronous subscription, the middleware will resend event messages to the subscriber immediately they are published on the topic, while on synchronous subscriptions, event messages are stored by the Platform until the subscriptor queries the topic for pending messages.
Fig. 6 shows graphically the proposed approach and Fig. 7 shows a UML sequence diagram to describe the interaction between participants.
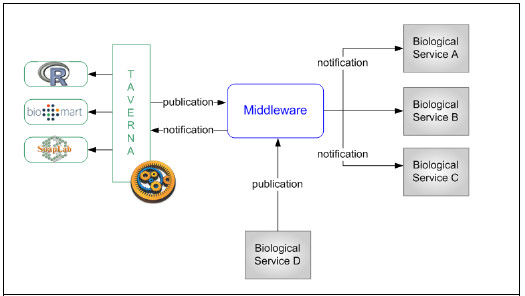
Figure 6: Middleware to notify events
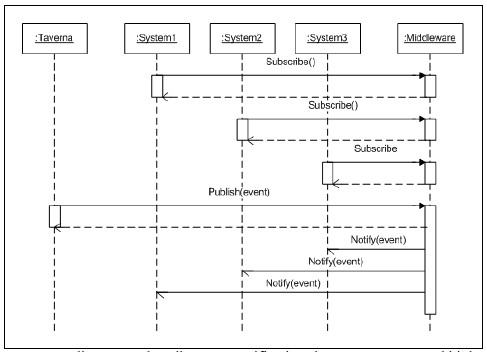
Figure 7: Sequence diagram to describe event notifications between Taverna and biological services.
The proposed solution based on an ESB Platform (Fig. 8) is designed using two design patterns: Event Driven Messaging and Protocol Bridging [25]. For each event of interest, there must be a topic configured in the ESB identified using a topicId. Each topic may have N Publisher and M Subscriptors, where each suscriptor may choose between two subscription models: asynchronous or synchronous subscription.
To publish an event on the Platform, the Publisher must create an event message with “business event data” and a topicId, and send it to the ESB Topic Endpoint. This component places the message into the topic according to the topicId and confirms its reception to the Publisher. After that, each subscribed service to this topic will receive a copy of the message according to its subscription type.
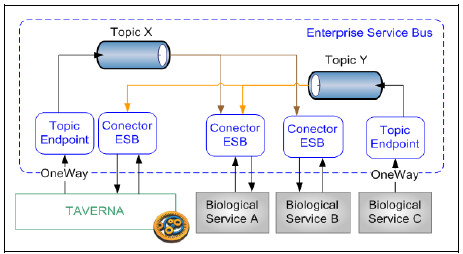
Figure 8: Events notification in the ESB
3.2.3 Message Transformation
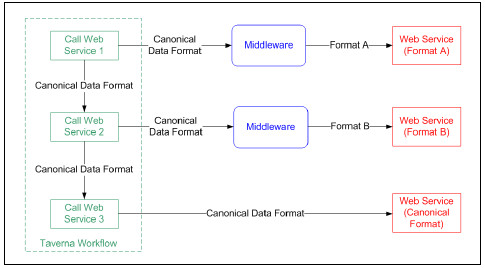
Scientific workflows are usually composed of different third-party services, where each service provider uses its own data format to model biological data. As a result, data format transformation must be solved repeatedly on each workflow. In [43] a survey of 415 workflows registered in myExperiment, found that 30% of the tasks involved in the experiment were due to format transformations, while just 22% were due to tasks involved in the invocation of Web Services. Therefore, the automation of these transformations generates significant productivity improvements in the development of bioinformatics experiments. The application of the Canonical Data Model pattern [25] is the preferred solution, but in cases where its usage is not possible, the middleware can perform message transformations to adapt the client’s requests to the service data format, and the service’s responses to the client’s expected data format. Fig. 9 shows graphically the proposed solution.
When instantiating the Platform with an ESB middleware (Fig. 10) the following design patterns are applied: Content-Based Routing, Canonical Data Model and Protocol Bridging [25]. The Plaform will automatically determine the format of the request messages and transform them to the expected data format. To reduce the amount of required processing, the Canonical Data Model design pattern may be applied to transform the incoming messages to a canonical data model and then from the canonical data model to the service data format. The Content Based Routing pattern is used to route incoming messages to the correct destinations. Native connectors are used to communicate the ESB with the biological services.
3.2.4 Service Composition
The low granularity of biological Web Services as well as the scarcity of coarse-grained Web Services adds complexity to the development of medium to big-size scientific workflows, as manual compositions must be developed each time the composed services are used. This scenario envisions the need to provide greater granularity of Web Services that allow an easier and fast development of biological experiments.
As in previous requirements, the use of middleware technologies is proposed to compose shim and biological services to develop more granular services, promoting Web Services composition reuse.
3.2.5 Service Policy Compliance
Nowadays, the leading biological web service providers (NCBI, EBI) provide their services without charge or fees to the whole scientific community, as long as they comply with their service policies. Failures to comply with these policies may result on severe penalties to consumers as show in Fig. 11 and 12.

Figure 11: NCBI service policy (http://www.ncbi.nlm.nih.gov/books/NBK25497/)

Figure 12: EBI service policy (http://www.ebi.ac.uk/Tools/webservices/help/interproscan)
Despite this service’s policies, Taverna provides little support for their compliance and enforcement. In this scenarios, scientists must develop custom tasks with advanced know how in informatics to comply with these policies. To overcome this issue, the Integration Platform provides new features to cope with this requirement to free the scientist of developing these low-level technical tasks and leverage policy compliance to the Platform as much as possible. Fig. 13 presents this proposal graphically.
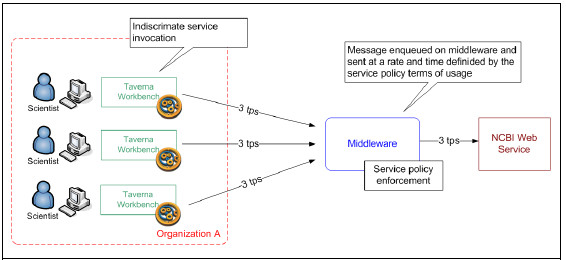
Figure 13: Middleware for service policy compliance
3.3 Context Usage
The Integration Platform can be applied in different bioinformatics laboratory contexts, which differ on their size and how their systems interact among each other, either consuming or providing services. Although every type of laboratory may have an Integration Platform providing the same functionalities, they may differ on the scope as well as on the supporting mechanisms. For example, asynchronous interactions features will be more limited in small-sized laboratories than in larger sized ones.
Therefore, prior to introducing the detailed specification of the Integration Platform, it’s necessary to define different types of laboratories, which characterize different application contexts. This typology is not intended to be neither a formal nor a complete classification, but as an intuitive categorization of usage contexts introduced in this work as any other prior categorization wasn’t found in the state of the art.
-
Laboratories type 1 (L1) use Taverna in a standalone mode, using only its native connectors to communicate with external biological services (Web Services, R connector, Biomart, etc.). These labs develop their experiments composing local and third-party services with Taverna as their main tool, and do not integrate with other services whose technology is not supported by Taverna.
-
Laboratories type 2 (L2) develop their experiments using local and third-party services compatible or not with Taverna’s native connectors. To cope with these incompatibilities they use middleware technologies to integrate with the external services. The middleware used by these labs is characterized to be peer-to-peer, solving each time specific technical integration issues where Message Queues and Web Services are the preferred choice. In this scenario the number of unsupported integrations is limited and small, and most of the integrations are done with Taverna’s native connectors.
-
Laboratories type 3 (L3) are characterized by large number of invocations to services not compliant with Taverna’s native connectors. The use of peer to peer middleware is no longer the most adequate solution and they require more sophisticated middleware Platforms to avoid its limitations and to improve the overall management and maintenance. An example of a middleware platform type is the Enterprise Service Bus (ESB).
-
Laboratories type 4 (L4) are service providers for laboratories of type 1, 2 or 3. One of the main requirements of these laboratories is to provide services with high levels of interoperability and accessibility. The type of middleware used by these laboratories will strongly depend on the quantity of services they provide. Laboratories with a small supply of services are characterized to use peer-to-peer middleware, while laboratories of large size with a great number of services, will use platform type middleware to avoid the limitations of point to point middleware.
-
Laboratories type 5 (L5) provide Platforms as a Service (PaaS). This type of lab does not provide nor consumes bioinformatics services. The provided services may be used by smaller sized laboratories with limited capacity to develop experiments based on the use of advanced infrastructure or large size middleware platforms. Amazon SWF is an example of such platforms.
As explained in Section 3.1 and Figure 2, in addition to the provided functionalities and the used middleware technologies, the context of usage constitutes a third dimension to characterize specific Integration Platforms.
3.4 Refining Process of the Specification
The process of refining the specification follows a top down approach starting with a first abstract level which is refined based on the three dimensions: usage contexts, functional requirements and middleware technologies. The combination of these dimensions enables to define a specific Integration Platform for a given usage context, satisfying a set of requirements and using certain middleware technologies.
It is possible to refine the model according to any of the three defined dimensions obtaining a first sublevel of refinement. For example, within the context of use dimension, it is possible to refine the model according to any of the five application scenarios: L1, L2, L3, L4 or L5. This dimension determines what usage the laboratory will give to the Platform. While type 2 (L2) labs will use the Platform to resolve the inconsistencies in the use of services, type 5 (L5) labs will use it to provide Platforms as a Service (PaaS). It is possible to further refine the model using one of the other two dimensions. For example, the middleware technologies dimension (Message Queues, Web Services and ESB). Using Web Services or Message Queues will provide point to point integrations following a given communication model, while using an ESB, introduces a bus-based integration of heterogeneous services and communication mechanisms.
However, there are restrictions on how the middleware dimension can be combined with the context of use dimension. For example, the combination (L2, ESB) is not acceptable, since it is impractical to use an ESB when the number of integrations is low. The same applies to the combination (L5, Web Services). It makes little sense to offer Web Services as a service, since its use is applied within an organization and not outside of it.
Finally, the model may be instantiated using the functional requirements dimension. Following with the previous example, the allowed values to refine the Platform are: asynchronous interactions, event notification, etc. This latter dimension provides a more detailed design of the Platform than the one provided by the middleware dimension, showing its behavior in a lower level design. Again, there are restrictions on the combination of dimensions, given that not in every case it is possible to combine the values of this dimension with the values of the middleware dimension. For example, message data format transformations are not directly supported by Web Service middleware.
4 Case example
After describing the Integration Platform and its features, this section presents a case example of the Integration Platform using a workflow-based experiment. The goal is to show the capabilities of the Platform through a realistic case taken from the myExperiment site.
4.1 Workflow-based experiment sample
The workflow-based experiment builds a phylogenetic tree from a protein sequence input data (http://www.myexperiment.org/workflows/3369.html). Fig. 14 shows a graphical view of the experiment and the workflow description provided by myExperiment site is the following:
This workflow accepts a protein sequence as input. This sequence is compared to others in the Uniprot database, using the NCBI BLAST Web Service from the EBI (WSDL), and the top 10 hits are returned (Nested workflow:EBI_NCBI_BLast). For each extracted hit, the Uniprot REST service returns the protein sequence in FASTA format. The workflow concatenates the 10 protein sequences and submits them as input to the EBI CLustalw service (Nested workflow EMBL_EBI_clustalw2_SOAP). These sequences are aligned and returned as results. Finally, the alignment is submitted to the EBI Clustalw_phylogeny service (Nested Workflow: clustalw_phylogeny), and a phylogenetic tree in phylip format is retuned. The workflow returned a list of protein sequences in FASTA format, a Clustalw alignment, and a phylogenetic tree.
Implementing this experiment in Taverna requires using nested workflows in order to apply the submitJob-getStatus-getResult pattern (described in section 3.2.1). As NCBI’s Web Services lack of asynchronous callback capabilities the experiment workflow has to perform a continuous checking of the job status before querying for the result. Scientists using Taverna have to implement this pattern each time they need to use this kind of service.
The workflow experiment case was adapted to better show the features of the Integration Platform. Instead of using a remote NCBI BLAST Web Service the experiment uses a local version of Blast Service, which can be downloaded and updated from the NCBI’s site. A laboratory may have a local version in order to avoid penalties due to overloading the public service and because some specific data quality processes are performed periodically. Fig. 15 shows the information flow between organizations and systems. In this case, this would be a Laboratory L3, as it has local services and is a mass service remote consumer.
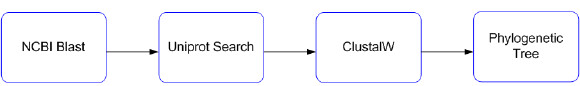
Figure 14: Experiment use case example
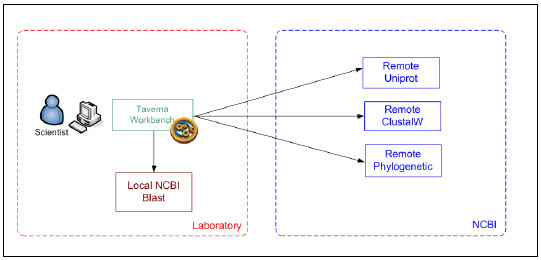
Figure 15: Workflow interactions and organizational boundaries
Finally, an extra requirement was introduced: every time the local BLAST database is updated (i. e. after data quality process or new NCBI data) it’s required to rerun the experiment in order to search for new results that can arise after the analysis of the new data. Nowadays, this must be done manually with Taverna.
4.2. Identified issues
This use case example poses three main issues for using Taverna as a standalone tool in large scale scenarios:
-
Workflow complexity: In order to achieve asynchronous interactions, NCBI developed their web services following the one-way request with status poll for completion interaction style (see Section 2.2.2). This adds a development complexity on every workflow that must use this type of services and promotes a misuse of computational resources.
-
Missing event based notifications: Re-running a workflow when new data is available must be a manual task, as Taverna does not have support for event based notifications that could react to.
-
No global service policy enforcement: To avoid penalties using NCBI Web Services (according to its terms of use), specific local development must be done in each workflow. In addition, a special coordination must be carried out by the scientific team as unexpected misuse can arise when using the same web service due to no central policy enforcer is available.
4.3. Integration Platform usage
In this use case example, the Integration Platform will provide callback asynchronous interactions, event based notifications, service composition and a global service policy enforcement support.
4.3.1 Asynchronous interactions
Fig. 16 shows the interactions between Taverna, the Integration Platform and the NCBI’s Web Services. Original asynchronous polling interactions where changed for asynchronous callbacks interactions. Every time Taverna must invoke Blast, ClustalW or the Pyhilogenetic web service, it must send a message to the Platform and wait for the callback response. The Platform is responsible for submitting the job, checking for the job status and querying for the results. This is done through a web service composition as it will be described later on in section 4.3.2.
For synchronous invocations like Uniprot web service, a web service proxy is defined in the Platform in order to resend request and responses synchronously.

Figure 16: Asynchronous interactions between Taverna, the Integration Platform and the Web Services
When the Platform receives a message, internally it is sent to a request message queue (Taverna Queue In) for further processing and an acknowledge response is sent to Taverna. A service composition is subscribed to the queue which takes care of all the interactions with the NCBI service. Responses generated by the service composition are sent to a response message queue (Taverna Queue Out) and later on sent to Taverna. Fig. 17 shows this behaviour.
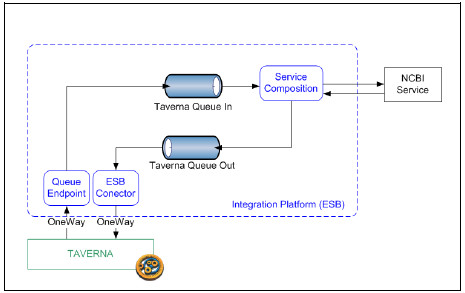
Figure 17: Integration Platform internal mechanisms
4.3.2 Service composition
In order to achieve callback asynchronous interactions, services compositions are needed to take care of the interactions between the Platform and the NCBI web services. Fig. 18 shows the service composition example usage for the NCBI services.

Figure 18: Service composition example usage
4.3.3 Event based notifications
To achieve event based notifications, it is necessary to define a new topic (Blast update topic) on the Integration Platform and add Taverna Runner to one of its subscribers. Every time an update is done to the local BLAST database, an updated notification is sent to the topic. When the Taverna Runner receives the notification it re runs the experiment defined in Taverna. Fig 19 shows this behaviour.

Figure 19: Event based notifications example
4.3.4 Service policy enforcement
In order to cope with this requirement, we restrict the number of subscribers of the Taverna Queue In message queue of the solution described in section 4.3.1. As every call to the NCBI service (even from different scientific teams) will be sent to the Integration Platform, restricting the number of subscribers allows to guarantee that no more than N jobs are submitted concurrently to NCBI. Besides further work must be done to accomplish full policy enforcement configuration, this is a step forward example to achieve this goal. Fig. 20 shows graphically this solution.
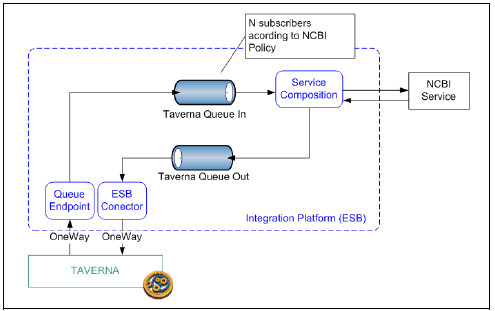
Figure 20: Service policy compliance
4.4 Evaluation
The presented a case example, a phylogentic tree analysis borrowed from the myExperiment site, enabled to show the Integration Platform capabilities in four areas: asynchronous interactions, service composition, event based notifications and service policy compliance.
We can conclude, firstly, that asynchronous interactions and the service composition capabilities provided by the Integration Platform reduced the development effort to develop bioinformatics experiments using NCBI’s services. This results in smaller tasks to accomplish the same experiment. In addition, event based notifications provide new ways of automation, looking forward to production chain experiments that could not be performed by Taverna standalone. Finally, service policy compliance capabilities provided better support to comply with NCBI’s terms of usage. Taverna’s capabilities are local by each experiment and require special coordination with team members in order to not misuse the services when running more than one experiment at the same time.
5 Formal Specification
The Integration Platform is formally specified using Event-B models [31] [32] [33]. Event-B allows the formal definition of the components of a system, the relations between them and the specification of their behaviour. The main objectives of using Event-B in this work are:
-
Provide precision to the system description: Event-B allows specifying precisely and unambiguously the behavior of the Platform.
-
The concept of virtual machine as system specification: Event-B allows the specification of a complete system, defining components, relationships and behavior.
-
Platform’s functionalities are event-based: The event-based nature of the Platform functionalities (notifications, asynchronism, etc.) motivates the use of this language as it is specific to model this type of systems.
-
Model refinements: Natively, Event-B provides the refinement concept, to model systems with different levels of abstraction, which is naturally applied to our needs on this work.
Therefore, the formal specification of the Platform will follow a hierarchical refinement defined by its levels of abstraction. In this paper, we model the first two levels of refinement of the specification. The UML-B tool was used to aid the specification development [44].
5.1 First Level of Refinement
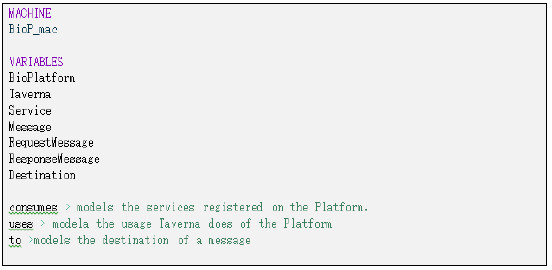
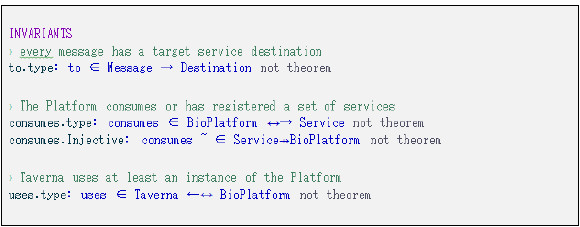
At the highest level, a "virtual machine" is specified for the first level of abstraction (Fig. 21). The components of the system are represented by variables (BioPlatform, Taverna, Service, etc.) as well as the relationships between them (consume, uses, etc.). Restrictions on components and relationships are presented in Fig. 22. The complete specification of this level can be found in [13].
5.2 Second Level of Refinement
In a second level of refinement (Fig. 23), the dimensions of middleware and functionalities are introduced, instantiated with an Enterprise Service Bus middleware and the "asynchronous interactions" functionality (section 3.2.1). The Machine defined in this level refines the Machine presented in Fig. 21, ensuring the solution is backward compatible and inherits its variables, invariants and events. The new variables introduced in this new second-level are variables related to subscriptions to the Platform (for receiving responses) and the necessary elements to specify its behavior. The BioPlatform element is used to model the ESB and its behavior.
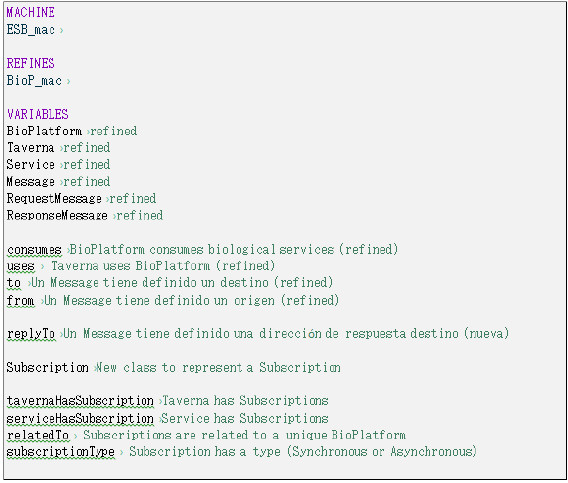
Figure 23: Second level refinement introducing an ESB middleware and the asynchronous interactions functionality
The restrictions defined in this level of refinement are: 1) "all received messages must specify a destination which must be one of the services registered on the Platform" and 2) "all request messages must provide a response address where to send the related responses". In Fig. 24 these restrictions are modeled using Event-B.

Figure 24: New invariants defined in the second level refinement
Fig. 25 and Fig. 26 provide a partial specification on Event-B of two events that reflect the behavior of the Platform when receiving a request and response message. In particular, the content-based router component of the Platform is modeled.
Every time a request message arrives to the Platform the processRequestMsg event is fired and the Platform will forward the message to the service specified in the to attribute. The event’s precondition is that the received message has specified its destination in the to message attribute and the service is registered in the Platform’s service registry (exists a “consumes” relationship between the Platform and the service). The pendingRequestMsgs relationship used by the event, models the received but not yet processed messages and the serviceRequests relationship, models the service’s received messages. Both relationships were specified in the first level of refinement and no further refinement was necessary. The processRequestMsg event refines the original event specified in the first level, as a new behavior is expected by this ESB-based platform instance.
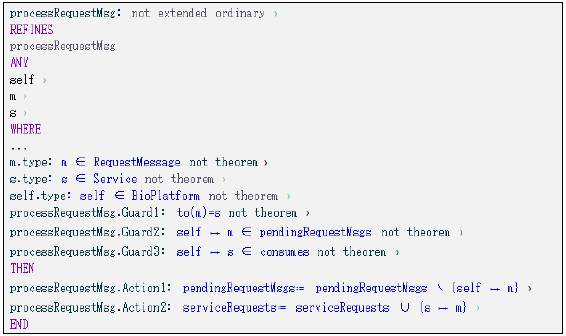
Figure 25: processResquestMsg event modeled using Event-B.
Every time a response message arrives to the Platform the processResponseMsg event is fired and will forward the message to an instance of Taverna assuming the following preconditions are met:
-
A Taverna instance is subscribed to the Platform using the asynchronous mode.
-
The Platform received a request message related to this response message and the relationship replyTo related to the request message has the Taverna instance as its value.
The first condition is modeled using two new relationships defined in this second level of refinement: tavernaHasSubscription and subscripionType. The first relationship represents the subscriptions Taverna has registered on the Platform, while the second one models the type of subscription it has (synchronous or asynchronous). The pendingResponseMsgs relationship models the response messages received by the Platform but not processed yet, while the tavernaReceives relationship models the response messages received by Taverna. Both relationships were specified in the first level of refinement and unchanged in this second level. The processResponseMsg event refines the original event specified in the first level, as a new behavior is expected by this ESB-based platform instance.
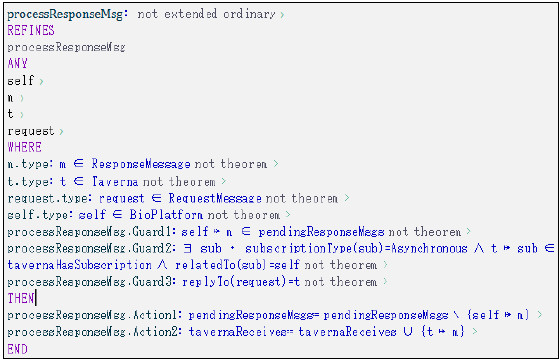
Figure 26: processResponseMsg modeled using Event-B
Fig. 27 presents the tavernaSubscribe event which models the process of registering a subscription for Taverna on the Platform to start receiving response messages. The subscription mode (synchronous or asynchronous) is defined by the subscription type (Subscription).
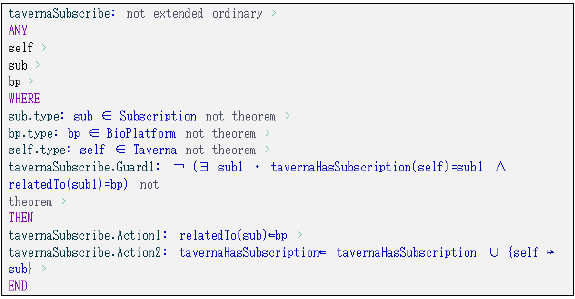
Figure 27: tavernaSubscribe event modeled using Event-B.
5.3 Specification Example
Let’s consider a simple example of the Platform’s behavior specified in the previous sections. This Platform is instanced using an ESB middleware, the “asynchronous interactions” functionality and a laboratory of type 3. In this use case, Taverna is owned by a laboratory of type 1, consumes biological services registered on the Integration Platform and has an asynchronous subscription to receive response messages.
Fig. 28 presents a sequence of events specified in Event-B representing the use case example. This is an illustrative representation and does not intend to be a formal specification of the events of the whole use case.
The first events that occur on the Platform are the registration of the biological services and the subscription of Taverna to receive response messages, which are modeled by the addService and tavernaSubscribe events. The process that models Taverna creating a request message and sending it to the Platform is modeled with the tavernaCreateMsgs and tavernaSendsMsgs events. The first event creates a message with the business data and the routing information to allow the Platform route the message to its final destination, and the second one, represents the act of sending the message to the Platform. Every request message is built with the to attribute having the destination service’s name and the replyTo attribute, to identify to which instance of Taverna the response must be sent to.
When the request message arrives to the Platform the event processRequestMsg is fired if the preconditions are met and the message is routed to the target service according to the message contents. After the service receives and processes the message (events serviceProcessMsg and serviceSendResponseMsg), it sends a response to the Platorm. The processResponseMsgs event is fired and the integration Platform will search for a related request message, read it’s replyTo attribute and identify the Taverna instance the message must be routed to. Before routing the message, the Platform will check this instance has a registered asynchronous subscription and only in that case, it will forward the response message to Taverna.
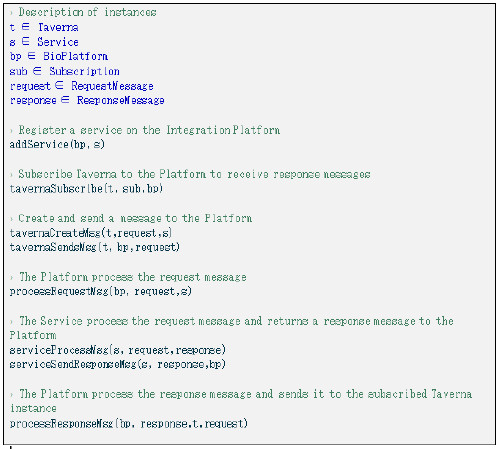
Figure 28: A specification example described using events modelled with Event-B.
6 Implementation
This article focuses on the ESB-based platform, which enables to implement the largest set of functionalities as well as to support the needs of the most complex application contexts (Laboratories type L3, L4, and L5). A description of implementations using the other middleware technologies (e.g. Web Services and Message Queues) can be found in [12].
6.1 ESB-based Implementation
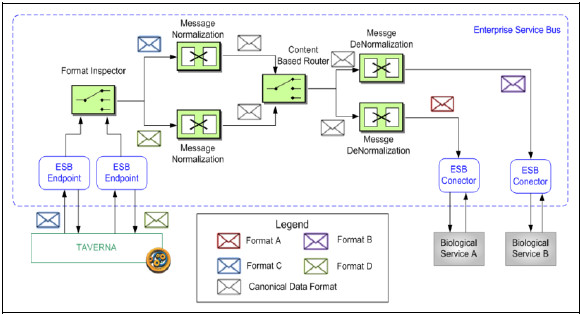
The ESB-based Integration Platform was implemented entirely using the JBoss ESB product (version 4.10), Java 6 Update 20 and Web Services from the NCBI (Blast, https://www.biocatalogue.org/services/1930 and ClustalW https://www.biocatalogue.org/services/3117), DDBJ (Blast https://www.biocatalogue.org/services/8 and ClustalW https://www.biocatalogue.org/services/7) and BCCS (Blast https://www.biocatalogue.org/services/49).
Fig. 29 shows the main components of the platform, identifying the ESB services with white rectangles. Each ESB service is composed of: 1) one or more providers to receive and respond message, represented as blue rectangles; 2) actions that perform the service implementation, represented as green (ESB native actions) and orange (custom actions) rectangles.
Most of the development was based on native ESB actions and a few specific custom actions where developed for those unsupported features by the ESB. The specification of the routing rules was necessary for each of the content-based routers and it was necessary to develop XSL transformations for those ESB services that required so.
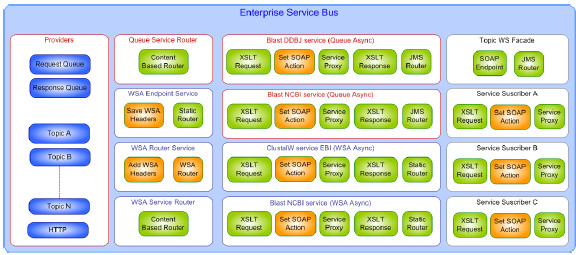
Figure 29: ESB-based Integration Platform components
6.1.1 Asynchronous Interactions
Asynchronous interactions based on message queues were implemented using native and custom actions of the ESB (Fig. 30). The native actions used were the Content Based Router and SOAPProxy, while a Jms-Provider was configured as the service provider (Fig. 31). It was necessary to develop a custom action for the management of specific http headers to integrate with SOAP Web Services, due to the SOAPProxy did not provide this feature.

Figure 30: ESB Service for asynchronous interactions
This prototype did not include the components in Taverna to send and receive messages from ESB queues using the asynchronous and synchronous subscription modes. In turn, a Java client was developed to simulate Taverna’s behaviour.
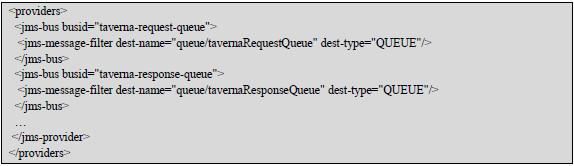
Figure 31: ESB JMS-Providers for asynchronous interactions
6.1.2 Events Notification
The events notification feature is fully implemented using ESB native components, particularly using the Jms-provider provider, which implements required features in the topic (Fig 32 and Fig 33).
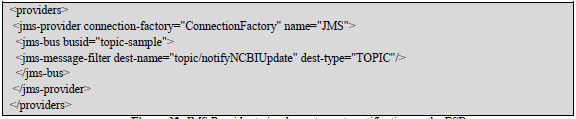
Figure 32: JMS Provider to implement events notification on the ESB

Figure 33: ESB-service simulating the real biological subscriptor
This prototype did not include the necessary components in Taverna to send and receive event messages from and to the ESB and neither real bioinformatics services subscribed to the event topics were used. These services were simulated using ESB-services subscribed using asynchronous subscriptions. Event messages where published on the Platform using a Java Client simulating the use of Taverna. The subscription process is done manually configuring the ESB services. APIs to perform this task programmatically were not implemented yet.
6.1.3 Message Transformation
This functionality was implemented with native and custom components of the ESB. Native components were the SOAPProxy and XsltAction actions and the http-provider. As in section 5.1.1, the development of a custom action was needed to manage specific http headers during the integration with biological Web Services because the SOAPProxy did not provide this feature.
To implement this prototype sample XSL transformations were developed as a proof of concept and validation of the proposed solution design, staying out of scope a complete implementation of the canonical data model. Transformations were only applied on the request messages, leaving response messages transformations to future work. Although this point is missing, the technical aspects to validate the solution were covered as the application of data transformation of the responses is similar as data transformation of the requests. Finally, transformations were applied to transform the message, but no changes were done on the published WSDL registered on the ESB to reflect the new format accepted by the service. The ESB service has the original WSDL published by the biological service but only accepts transformed messages. Changes to the ESB service WSDL is part of the future work.
6.2 Summary and Implementation Conclusions
The implementation allowed, first, to demonstrate the technical feasibility of the proposal, and secondly, to know in detail the technical issues involved while refining the Integration Platform on its third level of specification.
It also enabled to confirm that the selection of middleware technology to implement the Platform is strongly related to the type of laboratory. It is not reasonable to use ESB-based Platforms for laboratories of type 1 and 2 (L1 and L2 of section 3.3), nor the usage of Web Services for laboratories of type 3 (L3).
The cost/result varies according to the different choices. Experience after this work showed that using ESB middleware requires a high initial cost, both in learning and implementation for the initial configuration of the platform, which becomes marginal for each new service added to it. In turn, Web Services and message queues have short-term results given the shorter learning curve and implementation. In this type of middleware, the development cost is linear and constant for each service that you want to integrate to the platform. It’s also relevant to point out that laboratories which are not decided to use ESB as a first option may start with an implementation based on Web Services or Queues and to migrate afterwards to an ESB-based Platform, which does not constitute a complex task (using for example SOAPProxy or JMSRouter actions).
Some concrete development costs for integrating a service into the Platform using different middleware technologies are shown in Fig. 34 and Fig. 35. These figures were obtained from an engineering-degree project that involved the development of prototypes using Web Services and Messages Queues on one side, and an ESB on the other side. The results show the convenience of using Web Services and Message Queues-based Platforms for small size services environments, and ESB middleware type for medium to large size environments.
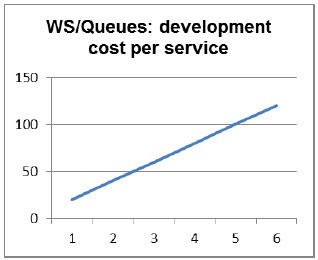
Figure 34: Platform development cost per service using Web Services or Message Queues as middleware.
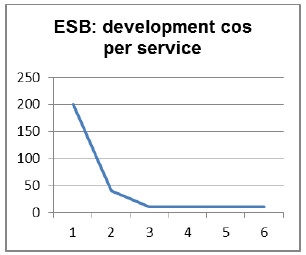
Figure 35: Platform development cost per service using ESB as middleware.
Furthermore, the implementation allowed us to know the state of the art on the products used, finding some gaps in some of them, particularly in the JBoss ESB. Specifically, it was necessary to develop custom actions to complement the use of native actions, due to shortcomings in some operations. For example, the SOAP Proxy did not specify the needed http header SoapAction to connect to a Web Service. Also, the lack of powerful development tools adds one step of complexity to development. The existing development tools were very poor (Eclipse plug-ins for JBoss Tools had few support for JBossESB features and some bugs where found) to the point that it was not used most of the functionality they provide. In contrast to this, Web services and message queues were found to be very mature technologies, with the expected behavior and supported enough by the development tools.
Finally, it was not possible in all cases to have a native integration between Taverna and the Middleware-based Platforms due to the lack of specific connectors for it. For example, Taverna lacks integration support with JMS Message Queues. However, implementation of such connectors would not be a problem because of extensible design Taverna provides for this purpose.
7 Conclusions and Future Work
This work addresses the issues of improving Bioinformatics laboratory collaboration and proposes a reference domain specific integration platform, which provides enhanced capabilities to implement distributed and service-based systems. This proposal aims at strengthening integration capabilities as well as reducing the complexity of application development by providing meaningful built-in mechanisms in the platform.
The implementation approach, based on enterprise middleware technologies (WS, ESB, etc.), shown to be capable of addressing the main requirements of providing advanced integration features and adequately connecting Taverna and other bioinformatics services. Nevertheless, functionalities like processing very large data sets require further work.
Although the proposed solution focuses on asynchronous interactions, event notifications and message transformation capabilities, it would also enable to improve process composition as well as the specification of service policy compliance.
The main contributions of this work consist of: (i) the context analysis and identification of relevant features to be provided in an advanced Bioinformatics integration platform, (ii) the proposed solution that can be applied to different laboratory contexts, (iii) the formal specification using Event-B which enables to perform a rigorous treatment of the platform features on its different refinement levels, and (iv) the implementation of prototypes that enabled to validate technologies and implementation approach.
The results obtained, based on an on-going research, enable to show the advantages and challenges of using middleware-based integration platforms to improve quality and scalability of in-silico bioinformatic experiments especially medium and large-scale ones. In any case, they show the feasibility of implementing a refinement-based approach, which would enable laboratories to incrementally adopt these solutions.
In addition, the work constitutes a step forward on carrying out a Platform as a Service (PaaS) approach for Bioinformatics. While the three-dimension framework (functionalities, scenarios, implementation platform) provides an extensible model, the design and implementation of specific mechanisms shows the feasibility of implementing such platforms and provides a highly useful practical experience.
Furthermore, the formal specification using Event-B, although it had a limited scope in this work, enables not only to provide a rigorous specification of this platform but also to incrementally build a specifications’ library for integration platform, and sets up the foundations for a formal treatment of this kind of systems.
This work opens several lines of future work, not only related to bioinformatics and the proposed platform, but also to the application in other domains. A first line consists of further implementation of in-silico experiments using the Integration Platform as well as to extend the provided functionalities and services, notably with service composition and service policy compliance management, and covering the transmission and processing of large data volumes and including connectors to languages often used in the Bioinformatic area (e.g. language R). A second line consists of developing a Platform as a Service (PaaS) approach for the proposed Platform, taking advantage of Cloud computing features to address Big Data scenarios related with the Next Generation Sequencing technologies. Third, future work would also extend the Event-B formal specification covering more features and including demonstrations of properties. This would enable to further conceptualize the integration platform and to provide a solid ground to extend functionalities.
Finally, conceptualizing Domain Specific integration platforms, beyond the Bioinformatics context, appears to be highly promising, as could enable to benefit from the conceptualization done in this work as well as parts of the technology platform to solve other integration requirements. The dimensions-based model may be replicated in different areas, parts of the Event-B specification could be reused and many of the ESB-based pattern implementation may be reused in other similar integration scenarios.
References
[1] R. Stevens, K. Glover, C. Greenhalgh, C. Jennings, S. Pearce, P. Li, M. Radenkovic, y A. Wipat, Performing in silico Experiments on the Grid: A Users’ Perspective. 2003.
[2] T. Kulikova, P. Aldebert, N. Althorpe, W. Baker, K. Bates, P. Browne, A. van den Broek, G. Cochrane, K. Duggan, R. Eberhardt, N. Faruque, M. Garcia-Pastor, N. Harte, C. Kanz, R. Leinonen, Q. Lin, V. Lombard, R. Lopez, R. Mancuso, M. McHale, F. Nardone, V. Silventoinen, P. Stoehr, G. Stoesser, M. A. Tuli, K. Tzouvara, R. Vaughan, D. Wu, W. Zhu, y R. Apweiler, «The EMBL Nucleotide Sequence Database», Nucleic Acids Res., vol. 32, n.o Database issue, pp. D27-30, ene. 2004.
[3] B. Boeckmann, A. Bairoch, R. Apweiler, M.-C. Blatter, A. Estreicher, E. Gasteiger, M. J. Martin, K. Michoud, C. O’Donovan, I. Phan, S. Pilbout, y M. Schneider, «The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003», Nucleic Acids Res., vol. 31, n.o 1, pp. 365-370, ene. 2003.
[4] S. F. Altschul, W. Gish, W. Miller, E. W. Myers, y D. J. Lipman, «Basic local alignment search tool», J. Mol. Biol., vol. 215, n.o 3, pp. 403-410, oct. 1990.
[5] J. D. Thompson, D. G. Higgins, y T. J. Gibson, «CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice», Nucleic Acids Res., vol. 22, n.o 22, pp. 4673-4680, nov. 1994.
[6] K. Wolstencroft, R. Haines, D. Fellows, A. Williams, D. Withers, S. Owen, S. Soiland-Reyes, I. Dunlop, A. Nenadic, P. Fisher, J. Bhagat, K. Belhajjame, F. Bacall, A. Hardisty, A. Nieva de la Hidalga, M. P. Balcazar Vargas, S. Sufi, y C. Goble, «The Taverna workflow suite: designing and executing workflows of Web Services on the desktop, web or in the cloud», Nucleic Acids Research, vol. 41, n.o W1, pp. W557–W561, jul. 2013.
[7] «Taverna in use | Taverna». [En línea]. Disponible en: http://www.taverna.org.uk/introduction/taverna-in-use/ . [Accedido: 03-nov-2014].
[8] G. Llambías y R. Ruggia, «Taverna: un ambiente para el desarrollo experimentos científicos», Pedeciba Informática, RT 10-11.
[9] G. Llambías, L. González, R. Ruggia, “Towards an Integration Platform for Bioinformatics Services”, Service-Oriented Computing – ICSOC 2013 Workshops, Vol. 8377, pp. 445-456, 2014.
[10] P. J. Hurd y C. J. Nelson, «Advantages of next-generation sequencing versus the microarray in epigenetic research», Brief Funct Genomic Proteomic, vol. 8, n.o 3, pp. 174-183, may 2009.
[11] M.-T. Schmidt, B. Hutchison, P. Lambros, R. Phippen, “The enterprise service bus: making service-oriented architecture real”, IBM Systems Journal, Vol. 44, nº 4, pp. 781–797, 2005.
[12] G. Wiederhold, “Mediators in the architecture of future information systems”, Computer, Vol. 25, nº 3, 38–49, 1992.
[13] G. Llambías, “Hacia una Plataforma de Integración de Servicios Bioinformáticos”, Tesis de Maestría. Pedeciba Informática. Uruguay, 2013.
[14] D. Hull, K. Wolstencroft, R. Stevens, C. Goble, M. R. Pocock, P. Li, y T. Oinn, «Taverna: a tool for building and running workflows of services», Nucleic Acids Res, vol. 34, n.o Web Server issue, pp. W729-W732, jul, 2006.
[15] B. Langmead, C. Trapnell, M. Pop, y S. L. Salzberg, «Ultrafast and memory-efficient alignment of short DNA sequences to the human genome», Genome Biology, vol. 10, n.o 3, p. R25, mar, 2009.
[16] A. Falk, T. Faber, J. Bannister, A. Chien, R. Grossman, y J. Leigh, «Transport protocols for high performance», Commun. ACM, vol. 46, n.o 11, pp. 42–49, nov. 2003.
[17] T. Disz, M. Kubal, R. Olson, R. Overbeek, y R. Stevens, «Challenges in large scale distributed computing: bioinformatics», en Challenges of Large Applications in Distributed Environments, 2005. CLADE 2005. Proceedings, 2005, pp. 57-65.
[18] L. D. Stein, «The case for cloud computing in genome informatics», Genome Biology, vol. 11, n.o 5, p. 207, may, 2010.
[19] I. Altintas, J. Wang, D. Crawl, y W. Li, «Challenges and approaches for distributed workflow-driven analysis of large-scale biological data: vision paper», Proceedings of the 2012 Joint EDBT/ICDT Workshops, New York, NY, USA, 2012, pp. 73–78.
[20] R. D. Stevens, A. J. Robinson, y C. A. Goble, «myGrid: personalised bioinformatics on the information grid», Bioinformatics, vol. 19, n.o suppl 1, pp. i302-i304, mar. 2003.
[21] S. Bechhofer, J. Ainsworth, J. Bhagat, I. Buchan, P. Couch, D. Cruickshank, M. Delderfield, I. Dunlop, M. Gamble, C. Goble, D. Michaelides, P. Missier, S. Owen, D. Newman, D. De Roure, y S. Sufi, «Why Linked Data is Not Enough for Scientists», ESCIENCE '10 Proceedings of the 2010 IEEE Sixth International Conference on e-Science, 2010, pp. 300-307.
[22] «Web Service Definition Language (WSDL)». [En línea]. Disponible en: http://www.w3.org/TR/wsdl . [Accedido: 03-nov-2014].
[23] D. A. Chappell, Enterprise service bus. Beijing; Cambridge: O’Reilly, 2004.
[24] C. Hérault, G. Thomas, y U. J. Fourier, «Mediation and Enterprise Service Bus: A position paper», en Proceedings of the First International Workshop on Mediation in Semantic Web Services (MEDIATE), 2005
[25] Erl, T.: SOA design patterns. Prentice Hall, Upper Saddle River, NJ (2009).
[26] G. Hohpe, B. Woolf, “Enterprise Integration Patterns: Designing, Building and Deploying Messaging Solutions”, Addison-Wesley Professional, 2003.
[27] Enterprise Connectivity Patterns: Implementing integration solutions with IBM’s Enterprise Service Bus products, http://www.ibm.com/developerworks/library/ws-enterpriseconnectivitypatterns/, [Accedido: 03-nov-2014].
[28] G. Edwards y N. Medvidovic, «A Methodology and Framework for Creating Domain-Specific Development Infrastructures», en 23rd IEEE/ACM International Conference on Automated Software Engineering, 2008, pp. 168-177.
[29] «IPF Overview - Open eHealth Integration Platform 2.x - Confluence». [En línea]. Disponible en: http://www.openehealth.org/display/ipf2/IPF+Overview. [Accedido: 03-nov-2014].
[30] B. Rienzi, L. González, y R. Ruggia, «Towards an ESB-Based Enterprise Integration Platform for Geospatial Web Services», GEOProcessing 2013, The Fifth International Conference on Advanced Geographic Information Systems, Applications, and Services, 2013, pp. 39-45.
[31] C. Métayer, J. R. Abrial, L. Voisin, Rigorous Open Development Environment for Complex Systems: Event B language. 2005.
[32] D. Yadav y M. Butler, «Rigorous Design of Fault-Tolerant Transactions for Replicated Database Systems using Event B», Rigorous Development of Complex Fault-Tolerant Systems, Lecture Notes in Computer Science, Springer , 2006, 2006, pp. 343-363.
[33] J. Bryans, J. Fitzgerald, A. Romanovsky, y A. Roth, «Formal Modelling and Analysis of Business Information Applications with Fault Tolerant Middleware», Proceedings of the 2009 14th IEEE International Conference on Engineering of Complex Computer Systems, Washington, DC, USA, 2009, pp. 68–77.
[34] S. Perera, D. Gannon, “Enabling Web Service extensions for scientific workflows”, WORKS ’06, Workshop on Workflows in Support of Large-Scale Science, 2006. pp. 1–10.
[35] T. Gunarathne, C. Herath, E. Chinthaka, S. Marru, “Experience with adapting a WS-BPEL runtime for eScience workflows”, Proceedings of the 5th Grid Computing Environments Workshop, 2009, pp. 7.
[36] Y. Huang, E. Slominski, C. Herath, D. Gannon, “Wsmessenger: A web services-based messaging system for service-oriented grid computing”, Sixth IEEE International Symposium on Cluster Computing and the Grid (CCGRID 06), vol 1, nº 8, pp 173, 2006.
[37] A. Alqaoud, I. Taylor, A. Jones, “Publish/subscribe as a model for scientific workflow interoperability”, Proceedings of the 4th Workshop on Workflows in Support of Large-Scale Science (WORKS’09), 2009, pp. 1.
[38] D. Gannon, M. Christie, S. Marru, S. Shirasuna, A. Slominski, “Programming Paradigms for Scientific Problem Solving Environments”, Gaffney, P.W. and Pool, J.C.T. (eds.) Grid-Based Problem Solving Environments. pp. 3–15. Springer US (2007).
[39] D. Hull, R. Stevens, P. Lord, C. Wroe, C. Goble, “Treating shimantic web syndrome with ontologies”, University, Milton Keynes, UK (2004).
[40] D. Zinn, S. Bowers, T. McPhillips, B. Ludäscher, “Scientific workflow design with data assembly lines”, Proceedings of the 4th Workshop on Workflows in Support of Large-Scale Science, 2009, pp. 14.
[41] M. Wilkinson, B Vandervalk, L. McCarthy, The Semantic Automated Discovery and Integration (SADI) Web service Design-Pattern, API and Reference Implementation, Journal of Biomedical Semantics, Vol. 2, No. 1. (2011)
[42] N. J. Loman y M. J. Pallen, «EntrezAJAX: direct web browser access to the Entrez Programming Utilities», Source Code Biol Med, vol. 5, p. 6, 2010.
[43] Wassink, I., Vet, P.E. van der, Wolstencroft, K., Neerincx, P.B.T., Roos, M., Rauwerda, H., Breit, T.M.: Analysing Scientific Workflows: Why Workflows Not Only Connect Web Services. Presented at the July (2009).
[44] Snook, C. and Butler, M., U2B, “A tool for translating UML-B models into B”, UML-B Specification for Proven Embedded Systems Design, chapter 6. Springer-Verlag (2004).