Servicios Personalizados
Revista
Articulo
Links relacionados
Compartir
CLEI Electronic Journal
versión On-line ISSN 0717-5000
CLEIej vol.17 no.1 Montevideo abr. 2014
Bioinformatics Performance Comparison of Many-core
Tile64 vs. Multi-core Intel Xeon
Myriam Kurtz
Dep. Informática, Campus Universitario, Universidad Nacional de Misiones, N3304 Misiones, Argentina
Francisco J. Esteban
Servicio de Informática, Edificio Ramón y Cajal, Campus Rabanales, Universidad de Córdoba, 14071 Córdoba, Spain
Pilar Hernández
Instituto de Agricultura Sostenible (IAS-CSIC), Alameda del Obispo s/n, 14080 Córdoba, Spain
Juan Antonio Caballero
Dep. Estadística, Campus Rabanales C2-20N, Universidad de Córdoba, 14071 Córdoba, Spain
Antonio Guevara
Dep. Lenguajes y Ciencias de la Computación, Campus de Teatinos, Universidad de Málaga, 29071 Málaga, Spain
Gabriel Dorado *
Dep. Bioquímica y Biología Molecular, Campus Rabanales C6-1-E17, Campus de Excelencia Internacional Agroalimentario, Universidad de Córdoba, 14071 Córdoba, Spain
and
Sergio Gálvez *
Dep. Lenguajes y Ciencias de la Computación, Campus de Teatinos, Universidad de Málaga, 29071 Málaga, Spain
Abstract
The performance of the many-core Tile64 versus the multi-core Xeon x86 architecture on bioinformatics has been compared. We have used the pairwise algorithm MC64-NW/SW that we have previously developed to align nucleic acid (DNA and RNA) and peptide (protein) sequences for the benchmarking, being an enhanced and parallel implementation of the Needleman-Wunsch and Smith-Waterman algorithms. We have ported the MC64-NW/SW (originally developed for the Tile64 processor), to the x86 architecture (Intel Xeon Quad Core and Intel i7 Quad Core processors) with excellent results. Hence, the evolution of the x86-based architectures towards coprocessors like the Xeon Phi should represent significant performance improvements for bioinformatics.
Resumen
Se ha realizado una comparación de rendimiento, entre el multiprocesador de sesenta y cuatro núcleos Tile64 y la arquitectura multinúcleo x86 Xeon, aplicado a algoritmos bioinformáticos. Hemos utilizado para la evaluación comparativa, el algoritmo de alineamiento de pares de secuencias biológicas MC64-NW/SW, que hemos desarrollado previamente para alinear ácidos nucleicos (ADN y ARN), y proteínas como el péptido. Este algoritmo utilizado en la experiencia, es una implementación mejorada del algoritmo de Needleman-Wunsch que sirve para realizar alineamientos globales de dos secuencias, y el algoritmo de Smith-Waterman que se utiliza para realizar alineamiento local de un par de secuencias biológicas.
Para realizar la comparación sobre arquitectura x86 (Intel Xeon Quad Core y los procesadores Quad Core Intel i7), hemos migrado el MC64-NW/SW, originalmente desarrollado para el procesador Tile64, con excelentes resultados.
Por lo tanto, estimamos que la evolución de las arquitecturas basadas en x86 hacia coprocesadores como el Xeon Phi, deben representar mejoras significativas en el rendimiento de la bioinformática.
Keywords: RISC, SoC, tiles, cores, multithreading, threads, processes, dynamic programming, cache memory.
Spanish Keywords:
núcleos, multi-hilo, hilos, procesos, programación dinámica, memoria cache.
Received 2013-09-08, Revised 2014-03-10, Accepted 2014-03-10
Corresponding author: Sergio Gálvez, Dep. Lenguajes y Ciencias de la Computación, Campus de Teatinos, Universidad de Málaga, 29071 Málaga, Spain; galvez@lcc.uma.es
* Authors who contributed to the project leadership.
1 Introduction
The current computer engineering and High-Performance Computing (HPC) are evolving very fast towards new Chip MultiProcessor (CMP) architectures, using different approaches. On the one hand, the General-Purpose Graphics Processing Units (GPGPU) allow integrating thousands of processing units whose real computing power can be only exploited if particular programming methodologies are followed. On the other hand, the many-core Central-Processing Units (CPU) like the Tile64 [ 1] and Xeon Phi [ 2] represent the microprocessor state-of-the-art. There are also other approaches that range from heterogeneous processors, like the Cell Broadband Engine (Cell BE), to architectures like the Epiphany, which is usually associated with Field-Programmable Gate Array (FPGA) cards, and the Transcede family of hardware architecture (formerly the PicoChip architecture) [ 3].
Since Tilera released the TilExpress-20G card, our research group has been studying the performance of its Tile64 processor for bioinformatics algorithms [ 4, 5]. The TilExpress-20G is a Peripheral Component Interconnect express (PCIe) card that can be used on any desktop Personal Computer (PC) running the Linux operating system, being controlled from such a host using a Command-Line Interface (CLI).
The Tile64 contains 64 Reduced Instruction Set Computing (RISC) tiles (cores) running at 866 MHz with three levels of cache memory, being interconnected through a high-bandwidth network called intelligent Mesh (iMesh). Each tile can execute an independent process, and they can communicate with each other through shared memory, or using message passing and channel based proprietary libraries. The card has 8 GB of Small Outline-Dual In-line Memory Modules (SO-DIMM) of Double Data Rate 2 (DDR2) Synchronous Dynamic Random-Access Memory (SDRAM) that can be used as shared and local memory, as well as Solid State Disk (SSD).
We have focused mainly on both pairwise and multiple sequence alignment algorithms. Every algorithm has been written in C, compiled in the host using a cross-compiler (tile-cc) and launched into the card using the PCIe bridge. Among these developments, we have published a variant of Needleman-Wunsch and Smith-Waterman algorithms [ 6], adapted to the Tile64 characteristics and named ManyCore64-NeedlemanWunsch/SmithWaterman (MC64-NW/SW) [ 7], which showed performance gains that ranged from 10x to 20x when compared with the fastest alternative. Currently, we are developing and testing other algorithms, like the ClustalW [ 5], and the Basic Local Alignment Search Tool (BLAST) specifically adapted to the Tile64 but, by now, MC64-NW/SW is the one with the best gain in performance. However, this RISC hardware technology from Tilera has not evolved significantly over the last four years, whereas other companies like Intel have developed advanced commercial products. That is the case of Intel Xeon Phi.
Therefore, we address the migration of the MC64-NW/SW from the Tile64 environment based on processes to the Xeon environment based on threads. This is the first stage in an eventual migration to the Xeon Phi coprocessor. We describe the main characteristics of the Needleman-Wunsch algorithm, as well as the improvements that we have introduced to take advantage of the Tile64 architecture. The main steps to migrate the C code from the process-oriented approach to the thread-oriented one are explained in this work. The performance results using several comparison charts are shown with the corresponding conclusions. Any aspects relative to the power consumption or operating voltages are out of the scope of this work, and therefore have not been considered.
2 Alignment of Sequences
One of the main research areas in bioinformatics is the comparison of nucleic acid (DNA and RNA) and peptide (protein) sequences. Indeed, aligning and analyzing such sequences allows to build dendrograms or phylogenetic trees, as well as to identify identities and differences among them, which can be useful to design molecular markers for breeding purposes and for identifying Protected Designations of Origin (PDO). A computational alignment of sequences (considered as strings) tries to match pairs of nucleotide or amino acid residues (characters) following a score function whose final value must be maximized. From a biological point of view, mismatches correspond to residue changes, whereas gaps correspond to insertions or deletions (indels). For instance, a variation in the DNA sequence that affects a single base is called a Single Nucleotide Polymorphism (SNP), as in the case of polymorphisms that we have used for quality control of the olive oil PDO [ 8].
The score function can be supported by a substitution matrix that provides a mark for each pair of characters: matches have a high score, whereas mismatches have a negative or low score. This leads to alignments computed by Dynamic Programming (DP) algorithms, like the Needleman-Wunsch (NW) and Smith-Waterman (SW). The NW provides a global alignment; i.e., an alignment where both sequences are considered in their entire lengths. On the other hand, the SW identifies the single similar-region in both sequences whose alignment provides the greater score; i.e., a local alignment. These algorithms have been improved in order to consider indels [ 6] and, hence, to obtain alignments with more biological relevance. Due to the high computing-power required by the DP aligners, many authors have implemented their own versions of the algorithms using specific hardware [ 9, 10].
Nonetheless, the DP algorithms are neither the only nor the most used approach to align sequences. Thus, the heuristics applies the biological knowledge to develop alignment algorithms that produce high-quality results in much less time under particular conditions, usually involving very similar sequences. These algorithms are mainly applied to very large sequences [ 11] or to large sets of long sequences [ 12]. Finally, other widespread bioinformatics tools, such as the BLAST [ 13] and ClustalW [ 14] apply both heuristic/probabilistic and DP methods. For example, the BLAST uses a heuristic to filter data, in order to decrease the amount of it on which to apply the SW algorithm.
2.1 A Parallel Approach: MC64-NW/SW
To estimate the potential of the Tile64 many-core processor, we implemented the algorithm MC64-NW/SW, which is a parallel version of the NW and SW algorithms.
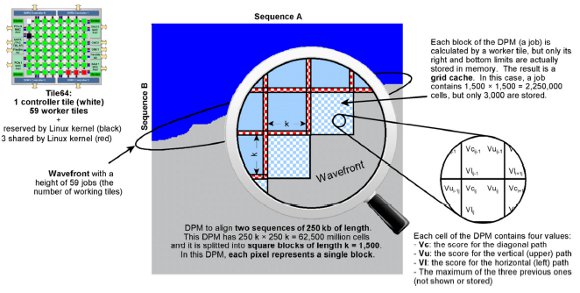
As the actual implementations of NW and SW are relatively similar, we will focus on NW only in this section. To align the sequences A and B with lengths n and m respectively, the original NW algorithm executes two phases: i) it creates a score Dynamic Programming Matrix (DPM) whose size is n×m; and ii) it traverses the DPM linearly to find out the actual alignment. Therefore, the first phase has a quadratic complexity both in time and memory space, whereas the second phase has a linear complexity. The MC64-NW integrates the FastLSA variant of NW [ 15] in order to overcome the huge memory requirements and, at the same time, to allow the alignment of long sequences. A FastLSA execution is represented in Figure 1.
Figure 1: Graphical description of the FastLSA parallel execution on the Tile64
To parallelize the first phase, the DPM is divided into submatrices or blocks of size k×k, so that the content of each block can be calculated by a tile: this is named a job. Once the top row and left column of a submatrix is available (obtained from the previous job completion or from the very first row and column initialization), a new job can be launched independently of any other running jobs. In turn, when a submatrix is completely calculated, only the last row and column are stored in the memory (eventually allowing a new job execution), discarding the submatrix content to free memory. The result is a grid cache; i.e., a sparse DPM where only one column and row are stored from every k. Therefore, the number of blocks/jobs into which the DPM is divided is [n/k] × [m/k] (see Figure 1), and thus, the total number of cells stored in the grid cache is 2k × [n/k] × [m/k]. Other details of the FastLSA are explained in [ 15], with Figure 1 showing the job distribution and, in turn, the content of every cell.
It is important to note that not every tile can work from the very beginning: when the algorithm starts, only the job at position (1, 1) can be computed by a single tile using the very first row and column of the previously initialized DPM. Afterwards, two jobs (2, 1) and (1, 2) can be computed in parallel by two tiles, using the results of the previous one and the initialization of the DPM. The conclusion is that the number of parallel jobs increases linearly and the DPM is created following a wavefront.
Thus, the full power of the system is achieved when the number of parallel jobs reaches the number of available tiles. The same happens at the end of the DPM generation. As a consequence, there is a tradeoff when choosing the best k value: the greater the k, the lower grid cache size, but the later that every tile is put to work, and vice versa.
The MC64-NW/SW uses a tile as the main controller to initialize the grid cache and the queue of jobs. In addition, the controller spawns the worker processes (one per tile) and controls the parallel execution, by sending jobs and receiving their results. As shown in the Figure 1, the number of actual tiles available to work is reduced from 64 to 59, since one is used as a controller, and four tiles are used for internal hardware tasks performed by the Linux kernel. Once the grid cache has been generated, the second phase starts at the bottom-right corner of the DPM and ends at its top-left corner, thus being named the backwards stage. To connect these two corners/cells, a linear path is followed throughout the DPM, depending on the content of the cells.
As the grid cache is stored, instead of the DPM, the submatrices where the path passes through must be recalculated. Thus, in this second phase, only a few submatrices must be recalculated: max (n, m) in the best case, and n + m – 1 in the worst scenario. This process is completely linear and it is performed by a single tile.
3 MC64-NW/SW in a x86 Multicore Architecture: MT-NW/SW
Migrating the MC64-NW/SW algorithm written in C to a x86 multi-core platform is not straightforward, mainly because the former uses the iLib proprietary library for communication, synchronization and process execution.
There are two main choices to substitute every iLib function call: message passing or shared memory. In general, it is not a good approach to execute each worker on a different process for a x86 muti-core architecture, because this may limit the shared memory capabilities. In addition, using the shared memory provides better results when there are not many concurrent accesses to the same memory, being this our case. Furthermore, this choice suits well with our future goal of reusing the new implementation for testing the Xeon Phi computing power: to take the most out of each Xeon Phi core, at least two threads must be executed to fulfill its hyper-threading capabilities [ 2]. Hence, the shared memory, threads and mutexes were selected. Three main changes have been made in the MC64-NW/SW to migrate it into the corresponding threads version (called MT-NW/SW, where MT stands for Many Threads):
Process spawn has been replaced by thread creation and initialization.
Message passing has been replaced by mutex access to shared memory.
Global variables have been encapsulated using an object-oriented-like programming methodology.
We have used the C language for the MT-NW/SW development, instead of any other advanced language (as C++) because, as stated above, we want to implement this same algorithm in other platforms like the Xeon Phi, and C has become a de facto standard for the emergent hardware architectures. The main drawback when adapting the algorithm from processes to threads has been to correctly manage the variables declared in the global scope. We used global variables in the MC64-NW/SW to store the progress of each job. This was found to be a good approach, because these variables are used throughout the C functions and an intensive usage of parameters is avoided. In addition, as the processes do not share the same runtime environment, there is no clash among the global variables stored in different processes (Tile64 allows sharing memory by means of passing messages with pointers to shared memory). Unfortunately, using threads implies that any global variable is visible by any thread, so a different methodology must be adopted. To solve this problem, we decided to simulate by hand the internals of an object-oriented compiler [ 16]. Basically, the state of a thread was encapsulated into a structure (like in an object), and a pointer to it was passed as a parameter to each function where the concept of this makes sense. Finally, the iLib functions have been replaced by new ones with the same functionality by means of mutexes. The resulting code of the MT-NW/SW is equivalent and as readable as that of the MC64-NW/SW.
4 Results
Several tests of the MC64-NW/SW running on 59 tiles [ 7] were compared against the MT-NW/SW using two different x86 environments:
1. Xeon Quad Core 2.0 GHz, 8 GB RAM DDR2 (quad-channel), running CentOS 5.3 (Dell Precision T5400). Two tests were carried out using 2 and 4 threads; thus, the relationship between a thread and a core was always one-to-one.
2. i7 3770 3.4 GHz (3.9 GHz in turbo mode), 8 GB RAM DDR3, running Ubuntu 12.2 (HP Pavilion p6-2307). Four tests were carried out using 2, 4, 6 and 8 threads. An additional test with a single thread was carried out as a reference. The i7 processor has four cores with two hyper-threading channels; therefore, using eight threads takes the most out of it, though its scalability was not linear when more than four threads are used.
With a simple calculation, it could be assumed that the top theoretical computing power of the Xeon Quad should be 4 × 2.0 GHz = 8.0 GHz, whereas that of the i7 3770 should be 8 × 3.9 GHz = 31.2 GHz. Likewise, the total theoretical computing power of the Tile64 should be 59 × 0.866 GHz = 51.09 GHz. Of course, this provides a measure for the power of the processors, but it does not take into account some other features, like the cache memories, bridges to the main memory, RAM specifications, etc. Therefore, it could be expected that, in the best scenario, the Tile64 should run at 6.39x when compared to the Xeon Quad, and at 1.64x when compared to the i7 3770.
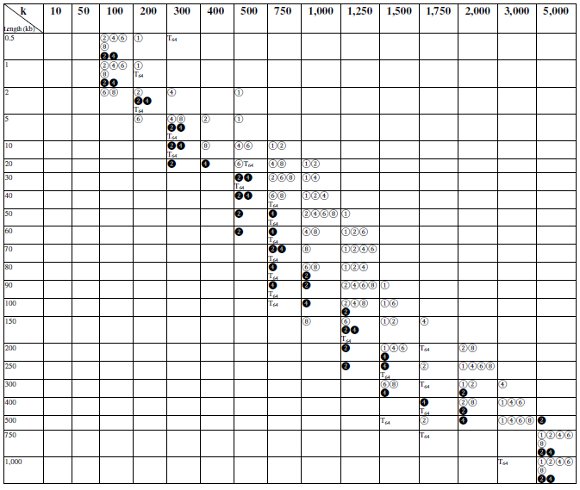
For this comparison, different k-values and sequences with different lengths were used for each test. The Table 1 shows the k-value that provided the fastest execution for each test and sequence length. The symbols ❷ and ❹ represent the executions on the Xeon Quad core environment with two and four threads, respectively, whereas the symbols ①, ② ④, ⑥ and ⑧ represent the executions on the i7 with 1, 2, 4, 6 and 8 threads, respectively. The symbol T64 represents the execution on the TilExpress-20G card using 59 tiles. For example, this table shows that using a Xeon Quad Core with only two threads to align sequences of 80 kb in length, the best k-value was 1,000, requiring 73,24 MB to store the grid cache [ 7]. From this table can be inferred that the best k-value was nearly the same when the MT-NW/SW is executed on the Xeon, no matter if two or four threads are used but, in the case of the i7 processor, as the number of threads increases, lower values of k usually produced better results.
The Tile64 behaved similarly to i7: it produced better results with low k values, because they allowed putting to work every tile in a shorter time.
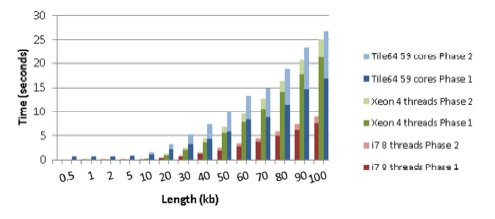
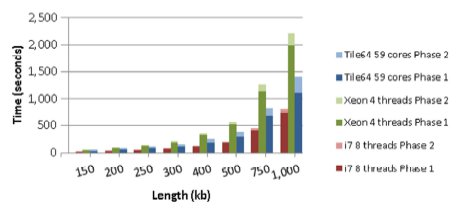
The best execution times are shown in Figures 2 and 3, where each group of three bars represents the time taken to align a pair of sequences with a particular length, using the three different environments. In turn, each bar is divided into two parts, being the dark one the time required to execute the first phase of the algorithm (whose complexity is quadratic), and the light one the time spent in the execution of the second phase (with linear complexity). The Figure 2 shows that the performance of the Tile64 environment was worse than those for any x86 environment when the two phases of the algorithm were considered. However, for sequences longer than 60 kb, the Tile64 executed the first phase faster than the Xeon Quad with four threads.
Clearly, the second phase took more time in the Tile64 because it was executed by a single tile whose computing power was near one third of that of a single core/thread in the Xeon Quad. On the other hand, with sequences longer than 100 kb, the Figure 3 shows that the Tile64 executed faster than the Xeon Quad, even when including the time consumed in the second phase, and the gain achieved using the many-core technology increased as the sequences to align became longer. Even more, the second phase took more and more time to be executed in the Xeon Quad because the first phase finished quicker with bigger k-values but, at the same time, the higher a k-value, the more time was spent in the second-phase execution.
Table 1: Best k-value for each executed test
Figure 2: Execution times for sequences with length from 0.5 kb to 100 kb
Figure 3: Execution times for sequences with length from 150 kb to 1,000 kb
The empirical comparisons between the Tile64 and the Xeon Quad when running the MC64-NW/SW and MT-NW/SW algorithms showed that the expected 6.39x gain was not achieved even with large sequences of 1,000 kb, where only an approximate 2x was obtained when considering exclusively the first phase. Therefore, the Tile64 behaved three times slower than theoretically expected. The main two reasons for this are:
1. The algorithm cannot take the most from every tile from the very beginning, but when the length of the wavefront matches the number of tiles. As more tiles are present, more time elapses until all of them are working.
2. The motherboards that integrate the x86 processors are more advanced and mature than that of the TilExpress-20G.
The assumption that the theoretical computing power of the Tile64 could be 1.64x when compared to the i7 3770 was nearly as optimistic as in the case of the Xeon Quad. Thus, the Tile64 executed the algorithm even slower than the i7 3770 in any test, with a performance that ranged from <50% (when aligning sequences with a length lower than 100 kb), up to 65% (when aligning the largest pair of sequences with a length of 1,000 kb). Actually, in such a case, the Tile64 was not three times slower than expected (as in the comparison against the Xeon Quad) because the factor 1.64x calculated above assumed an ideal one-to-one relationship between threads and cores in the i7 3770, which was not the case (this processor has only four cores with hyper-threading).
5 Conclusions and Future Perspectives
The analysis of the tests carried out in this work shows a clear advantage of the new Complex Instruction Set Computing (CISC) technology over the tested RISC one. This is evident even in the case of MC64-NW/SW, an algorithm that behaves extremely well in Tile64 in terms of speed. On the other hand, it should be taken into account the cheaper i7 3770 platform versus the expensive TilExpress-20G one.
However, from a retrospective point of view, the algorithms developed for the Tile64 processor performed very fast when compared with other technologies in the past. Therefore, our next work is to test the MT-NW/SW on an Intel Xeon Phi-coprocessor PCIe card, integrating 60 cores with four hyper-threading channels each and 8 GB of DDR5. With such a work we want to find out if the Xeon Phi technology boosts the performance –when compared to other current technologies– as well as Tile64 did six years ago. To do this, we need to proceed with another migration in order to take advantage of the particular characteristics of this new coprocessor. In particular, we are developing a variant to take the most out of the Xeon vectorization capabilities and, at the same time, we are applying a new approach to launch immediately the four threads of each core, just when it starts a job. Depending on the performance achieved, more accurate algorithms could be developed in order to achieve, for example, contig assemblies with a higher quality when dealing with de novo sequencing, BLAST executions with parameter values that allow more in-depth searching, more accurate dendrograms, etc.
Acknowledgements
We are grateful to Tilera for providing hardware and software tools http://www.tilera.com/. This work was supported by Ministerio de Economía y Competitividad (MINECO grants AGL2010-17316 and BIO2011-15237-E) and Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (MINECO and INIA RF2012-00002-C02-02); Consejería de Agricultura y Pesca (041/C/2007, 75/C/2009 and 56/C/2010) and Consejería de Economía, Innovación y Ciencia (AGR-7322) of Junta de Andalucía; Grupo PAI (AGR-248); and Universidad de Córdoba (Ayuda a Grupos), Spain.
References
[ 1] S. Bell, B. Edwards, J. Amann, R. Conlin, K. Joyce, V. Leung, J. MacKay, M. Reif, L. Bao, J. Brown, M. Mattina, C.-C. Miao, C. Ramey, D. Wentzlaff, W. Anderson, E. Berger, N. Fairbanks, D. Khan, F. Montenegro, J. Stickney, and J. Zook, TILE64 - Processor: A 64-Core SoC with Mesh Interconnect, presented at the ISSCC 2008, 2008.
[ 2] Parallel Programming and Optimization with Intel Xeon Phi Coprocessors. Colfax International, 2013.
[ 3] A. Vajda, Programming Many-Core Chips. New York: Springer, 2011.
[ 4] S. Gálvez, D. Díaz, P. Hernández, F. J. Esteban, J. A. Caballero, and G. Dorado, Next-Generation Bioinformatics: Using Many-Core Processor Architecture to Develop a Web Service for Sequence Alignment, Bioinformatics, vol. 26, no. 5, pp. 683–686, 2010.
[ 5] F. J. Esteban, D. Díaz, P. Hernández, J. A. Caballero, G. Dorado, and S. Gálvez, Direct approaches to exploit many-core architecture in bioinformatics, Future Gener. Comput. Syst., vol. 29, no. 1, pp. 15–26, 2013.
[ 6] O. Gotoh, An improved algorithm for matching biological sequences, J. Mol. Biol., vol. 162, no. 3, pp. 705–8, 1982.
[ 7] D. Díaz, F. J. Esteban, P. Hernández, J. A. Caballero, G. Dorado, and S. Gálvez, Parallelizing and optimizing a bioinformatics pairwise sequence alignment algorithm for many-core architecture, Parallel Comput., vol. 37, no. 4–5, pp. 244–259, 2011.
[ 8] A. Castillo, P. Pascual, E. Rodríguez, D. Díaz, M. G. Claros, J. Falgueras, S. Gálvez, G. Dorado, and P. Hernández, Genomic approaches for olive oil quality control, in Plant Genomics European Meetings, Tenerife, Spain, 2007.
[ 9] Y. Liu, B. Schmidt, and D. L. Maskell, CUDASW++2.0: enhanced Smith-Waterman protein database search on CUDA-enabled GPUs based on SIMT and virtualized SIMD abstractions, BMC Res. Notes, vol. 3, p. 93, 2010.
[ 10] M. Farrar, Striped Smith-Waterman speeds database searches six times over other SIMD implementations, Bioinformatics, vol. 23, no. 2, pp. 156–61, 2007.
[ 11] A. L. Delcher, S. Kasif, R. D. Fleischmann, J. Peterson, O. White, and S. L. Salzberg, Alignment of whole genomes, Nucleic Acids Res., vol. 27, no. 11, pp. 2369–2376, 1999.
[ 12] M. Brudno, C. Do, G. Cooper, M. F. Kim, E. Davydov, E. D. Green, A. Sidow, and S. Batzoglou, LAGAN and multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA, Genome Res., vol. 13, pp. 721 – 731, 2003.
[ 13] S. F. Altschul, W. Gish, W. Miller, E.-M. Myers, and D. J. Lipman, Basic local alignment search tool, J Mol Biol, vol. 215, pp. 403–410, 1990.
[ 14] M. A. Larkin, G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins, Clustal W and Clustal X version 2.0, Bioinformatics, vol. 23, no. 21, pp. 2947–2948, 2007.
[ 15] A. Driga, P. Lu, J. Schaeffer, D. Szafron, K. Charter, and I. Parsons, FastLSA: A Fast, Linear-Space, Parallel and Sequential Algorithm for Sequence Alignment, Algorithmica, vol. 45, no. 3, pp. 337–375, 2006.
[ 16] M. Siff and T. Reps, Program generalization for software reuse: from C to C++, in Proceedings of the 4th ACM SIGSOFT symposium on Foundations of software engineering, New York, NY, USA, 1996, pp. 135–146.