Introduction
The dairy industry in Uruguay comprises an essential sociocultural and economic activity. It has grown steadily in recent decades and represents the second most relevant activity after the meat supply chain activity (Ministerio de Ganadería, Agricultura y Pesca, 2021). Uruguay is one of the largest dairy producers and is one of Latin America’s highest per capita consumption countries of dairy products (Fariña and Chilibroste, 2019). The practice of intensive production systems improves productivity. It increases economic profits, but the confinement of animals, associated stress, and inadequate and insufficient colostrum consumption between other management`s risk factors promote a steady spread of infectious disease (Lorenz et al., 2011). Colostrum is the first postpartum secretion from the mammary gland, and its ingestion by the newborn calf is vital due to its high content of immunoglobulins (Godden, 2008). Passive immune transfer (PIT) of maternal immunoglobulins to the calf depends on factors such as the quantity and quality of colostrum supplied and the time it is fed to the calf (Godden et al., 2019). High-quality colostrum intake constitutes the primary source of antibodies during the first weeks of life (McGuirk and Collins, 2004) and correlates with good tracers of health and survival of calves of dairy breeds during the suckling stage (Heinrichs et al., 2020;National Dairy Heifer Evaluation Project, 1993; Schild, 2017).
Neonatal calf diarrhea (NCD) is a severe infectious disease that challenges livestock industries worldwide (Lorenz et al., 2011). In South America, NCD prevalence and mortality rates can reach almost 60 % and 20 %, respectively (Margueritte et al., 2007). In Uruguay, calf mortality rates in dairy farms, associated with infectious diseases, are high, and mainly occur in the first 21 days of calf life (Schild, 2017). Among the most frequent pathogens associated with NCD we found Cryptosporidium spp., Rotavirus, and pathogenic Escherichia coli (Caffarena et al., 2021; Schild, 2017).
The adhesion of E. coli to the intestinal mucosa is a key step in the infectious process. CS31A is a plasmid-encoded adhesin, immunologically and structurally related to the F5 fimbriae found in bovine isolates, and also linked with F17 and P fimbriae and Cnf1 toxins Girardeau, 1988 Le Bouguénec and Bertin, 1999. The non-fimbrial adhesin CS31A from E. coli, is associated with NCD. The clpG gene, which codifies for the structural subunit of the adhesin (ClpG), has one of the highest prevalence in calves feces in the region (Mercado et al., 2003; Umpiérrez et al., 2016, 2021), and it is frequently detected in diarrheic and septicemic calves around the world (Kolenda et al., 2015; y Nagy and Fekete, 1999).
Vaccination against NCD improves the colostrum immunoglobulin composition in pregnant cows. Its correct consumption ensures the transmission of antibodies to the calf after birth and should prevent the occurrence of diseases (Nagy and Fekete, 2005). There are several vaccines against NCD on the market, which frequently contain E. coli bacterin or purified E. coli fimbriae together or not with toxoid, as well as different viral antigens (Nagy and Fekete, 2005). Since cross-protection to E. coli fimbrial antigens through colostrum consumption is not possible, vaccines whose composition is adjusted to the outbreaks and local-circulating strains are required (Maier et al., 2022). Among bacterial antigens, some vaccines contain bacterin from E. coli CS31A+ strains. However, no vaccine evaluation studies have been carried out on the presence of specific antibodies against CS31A nor the passive transfer of this antigen to the calf through colostrum yet.
This work aimed to determine the level of IgG anti-ClpG antibodies in the colostrum of dairy cows without vaccination and vaccinated against NCD in Uruguay.
Materials and Methods
Bacterial strains and plasmids
The strain Escherichia coli 3.4A (clpG+) was isolated from the feces of a calf with NCD symptoms in Uruguay in 2012 (Umpiérrez et al., 2016). E. coli XL-1 Blue was used to transform and stabilize the expression plasmid (Bullock et al., 1987), while E. coli BL21 (λDE3) was used to express ClpG (Studier and Moffatt, 1986). The latter is lysogenic for the DE3 phage, which has the gene that codes for T7 RNA polymerase under the Lac UV5 promoter (inducible by isopropyl-β-D-thiogalactopyranose - IPTG). Plasmid pET-21a (+), resistant to Ampicillin, has the T7 promoter, and was used to express the protein (Studier and Moffatt, 1986).
Isolates were grown in Luria Bertani Agar plates (LB) at 37 °C for 18-24 h. In cases where it was required to select E. coli colonies that presented the plasmid pET-21a(+)-clpG, Ampicillin was added at a final concentration of 100 µg/mL.
Amplification of the clpG gene by PCR
ClpG was amplified and cloned into the expression plasmid pET-21a(+) (Bahrani and Mobley, 1994) using E. coli 3.4A genomic DNA as a template for the amplification (Umpiérrez et al., 2016). The E. coli 3.4A genomic DNA was extracted with the commercial kit GenElute Bacterial Genomic DNA kit (Sigma) according to the supplier’s instructions, and its concentration was quantified with Nanodrop (Thermo).
The primers ClpGr-F 5´-GGAATTTATCATATGAAAAAGACTCTGATTGC-3´ and ClpGr-R 5´CGCCCTTAAGCTTAATGGTGATGGTGATGGTGGTAAAGGTTGCTTCAATAGTC-3´ for the amplification of clpG gene were designed according to published sequences (GenBank accession number M55389.1), and the restriction sites for NdeI and HindIII enzymes (New England, BioLabs) were incorporated into their sequences in order to amplify the gene. Also, a histidine tag was included in the reverse primer ClpGr-R for subsequent protein purification.
PCR was performed in a 2720 Thermal Cycler (Applied Biosystems) thermocycler, using 0.1 mM dNTPs, 0.32 μM of each primer, 2 μL of DNA, 1.5 mM of MgCl2, 1X of buffer, and 1U Taq DNA polymerase (Invitrogen, Waltham, USA). Briefly, the cycling program comprised an initial activation step at 94 °C for 3 minutes, 30 cycles of amplification (denaturation at 94 °C for 1 minute, annealing for 1 minute at 48 °C followed by an extension at 72 °C for 1 minute), a final extension step at 72 °C for 5 minutes, and chilling at 4 °C. The clpG PCR product was digested with NdeI and HindIII, according to the protocol of the enzymes (New England, BioLabs) and ligated into pET-21a(+), which was previously digested with the same enzymes. The ligation mixture pET-ClpG was used to transform E. coli XL-1Blue according to standard procedures (Sambrook et al., 1989). Finally, the plasmid pET-ClpG was moved from E. coli XL-1Blue into E. coli BL21 (λDE3) via transformation.
Recombinant ClpG overexpression in vitro
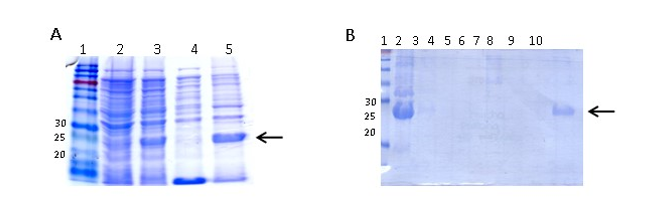
The expression of recombinant ClpG (ClpGr) was performed according to the procedure of Sambrook (Sambrook et al., 1989). Briefly, 200 mL of LB / Amp broth ;100 µg/mL; was inoculated with an isolated colony of E. coli BL21 (λDE3) (pET-ClpG), and the medium was incubated at 37 °C with vigorous shaking. Once the culture reached an OD600 between 0.4 and 0.8 nm, the expression of the protein was induced by adding IPTG to a final concentration of 1 mM. The culture was incubated with vigorous shaking for 3 h at 37 °C and centrifuged at 10,000 rpm and 4 °C for 15 min. The pellet containing the cells was stored at -20 °C for the subsequent purification of ClpGr. The expression of the recombinant protein was monitored by SDS-PAGE.
ClpGr purification and quantification
ClpG purification was carried out according to the purification protocol described by Qiagen under denaturing conditions. For the extraction of inclusion bodies (IB), cells previously induced with IPTG were resuspended in 40 mL buffer (50 mM Tris, 0.5 M NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF) and lysozyme (1 mg/mL) and were incubated on ice for 30 min. DNAse (1 mg/mL) was then added and incubated for 30 min on ice. The cells were sonicated intermittently on ice to complete the lysis at 200-300 W for 5 min (X6). When a clear lysate was obtained, 2 % Triton X100 was added and incubated for 30 min on ice. The IB in the pellet were obtained by centrifugation at 15,000 rpm at 4 °C for 30 min. Subsequently, they were washed in 3 successive centrifugations in 40 mL of buffer (50 mM Tris, 0.5 M NaCl), centrifuging at 15,000 rpm for 15 min at 4 °C. Finally, the IB were resuspended in 10 mL of buffer A (8 M Urea, 10 mM Tris, 0.1 M NaH2PO4, pH = 8). The lysate obtained was incubated for 1 h at RT and then centrifuged at 10,000 rpm and 4 °C for 15 min. To the obtained supernatant, 5 mL of 50 % Ni-NTA (Qiagen) balanced resin wasadded and the resin and lysate mixture was incubated for 45 min at RT, with shaking. Once the column was left in static conditions, the resin decanted and was washed with buffer A with an outlet flow of approximately 10-15 mL/h, until the absorbance at 280 nm was less than 0.01. Then, the column was washed with buffer C (8 M Urea, 10 mM Tris, 0.1 M NaH2PO4, pH = 6.3), and again the Abs280 was checked to be less than 0.01. Subsequently, the nickel-bound protein was eluted from the column by adding buffer C and Imidazole to a final concentration of 500 mM. The fractions were collected in several tubes, of which the first contained 15 mL of eluate and the next 5 mL each. Finally, the presence of the protein in each fraction was assessed by 15 % SDS-PAGE. Those fractions where the protein was found were dialyzed against distilled water at 4 °C for 72 h using a dialysis membrane that retains proteins whose molecular weight (MW) is greater than 12 kDa (Sigma). The dialyzed protein was stored in aliquots at -20 °C until use.
Sample collection: colostrum from vaccinated and unvaccinated dairy cows
Colostrum from 30 milking cows was collected from three dairy farms between 2019 and 2020, in 3 different geographical location in Uruguay (Table 1).
Table 1: Origin of the dairy herds, year of colostrum collect and breed of vaccinated and unvaccinated cows
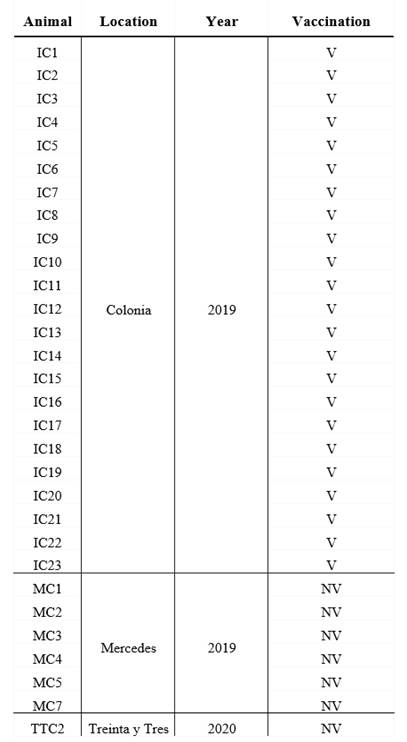
V: vaccinated, NV: non-vaccinated. Colonia, Mercedes and Treinta y Tres are 3 different geographical locations in Uruguay.
All cows were Holstein-Friesian between the second and the fourth lactation and were healthy at the moment of sample collection. A volume of 200 mL of each colostrum was collected in sterile bottles within the first day after calves` birth, and chilled before arriving at the laboratory. All colostrum samples were fractionated in aliquots and maintained at -20 °C until use.
The colostrum form unvaccinated cows (n=7) came from small dairy farms where no vaccines are administered to pregnant cows, whereas the vaccianted cows (n=23) came from a controlled farm and recieved 2 doses, 4 weeks apart, of a vaccine during the pregnancy (the second dose was administered 5 weeks before the expected date of delivery) (Table 1). Vaccination of pregnant cows to prevent infectious deseases in neonates is a widespread strategy in Uruguay, which made it difficult to obtain colostrum from unvaccinated cows. Vaccine components include chemically inactivated cultures of bovine Rotavirus G6P5, bovine Rotavirus G10P11, bovine Coronavirus, E. coli J5, E. coli K99, E. coli F17/CS31A and Clostridium perfringens type C adsorbed in aluminum hydroxide. The route of administration and the immunization protocol used were those established by the vaccine manufacturer.
Colostrum composition
Colostrum components were analyzed using a portable ultrasonic milk analyzer (Lactoscan SPL, Milcotronic, Bulgaria) previously calibrated with standardized methods (International Organization for Standardization (ISO), 2013). Each colostrum was processed by duplicate and the assessment included fat, protein and lactose.
Indirect ELISA to determine colostrum-specific antibodies against ClpG
The presence of specific IgG colostrum antibodies was determined by an indirect enzyme-linked immunosorbent assay (ELISA) set-up in this work. Plates were coated with 3 µg/mL of ClpGr suspended in carbonate buffer (pH=9.6) per well and incubated overnight at 4 °C. Colostrum samples were diluted 1:150 and incubated for 90 min at 37 °C. All colostrums were evaluated in triplicate. Bound antibodies were detected by adding peroxidase-labeled anti-bovine IgG (whole molecule) diluted 1:500 (KPL); plates were incubated for 30 min at 37 °C. After washing, o-phenylenediamine dihydrochloride substrate tablets (Thermo) dissolved in 0.2M Na₂HPO₄ and 0.1M citric acid buffer was added. Plates were incubated for 5 min at room temperature, and the reaction was stopped by adding H2SO4. Finally, absorbance at 490 nm was measured with an ELISA microplate reader (VarioskanFlash, Skanlt Software 2.4.3 RE). For the interpretation of ClpG antibody levels, cut-offs were established according to Wright et al. 1997, who suggest using a positivity range between 40-65 % of the strong positive (positive sample) to establish the weak positive value and 0-10 % of the strong positive for the negative of the technique.
Statistical analysis
The Kruskal-Wallis test for equal medians (PAST 4.03) was employed to determine possible differences between groups of animals in the content of fat, protein and lactose of the colostrum (p-values < 0.05 were considered significant).
The differences between absorbance values among vaccinated and unvaccinated groups of cows were studied by means of linear and log linear model using likelihood ratio test (LRT) using a general model: a is a common intercept, bi is the coefficient for the groups, (i) mean, ε are the residuals which follows a Normal (N) distribution, with mean 0 and variance e i σ 2 , where e i is a group coefficient for variance. The models were fitted using “gls” function from package {nlme} with three different structures:
i) mean varying for each group and equal variance (bi, e i =1),
ii) equal mean and different variances for each group (bi=0, e i ),
iii) different mean and variance for each group (bi, e i ).
To evaluate variance differences among groups of cows, the log-likelihood ratio between models i and iii and between models ii and iii to evaluate differences in mean values among groups. Under the null hypothesis, the log-likelihood ratio follows a Chi-square distribution, Figure 1.
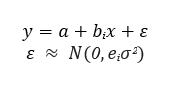
Results
Expression and purification of recombinant-ClpG protein
The amplification product from the E. coli 3.4A strain was subcloned into the expression plasmid pET21a(+) and subsequently incorporated by transformation into the E. coli BL21(λDE3) strain. The insert containing the clpG gene in the plasmid was confirmed by plasmid purification and enzymatic digestion (not shown).
ClpGr was expressed under denaturing conditions after the induction of an E. coli BL21(λDE3)-ClpGr LB culture with IPTG 1mM. Induction of ClpGr expression was confirmed by running total protein extracts before (t0) and after (tf) IPTG addition on polyacrylamide gels under denaturing conditions (Supplementary Fig 1). The recombinant protein ClpGr was purified using Agarose-Nickel affinity columns with 6M urea, and dialyzed against saline prior to its utilization.
Colostrum composition and presence of anti-ClpG antibodies
In this work, colostrum from 30 milking cows in Uruguay were evaluated for its fat, lactose and protein composition and for the presence of IgG anti-ClpG CS31A fimbriae (7 non-vaccinated cows and 23 vaccinated cows). For Lactoscan analyses, the volume of colostrum samples from 2 vaccinated cows (IC15 and IC22 animals) were insufficient and were not considered in the compositional screening. For the ELISA assay, all the samples were employed.
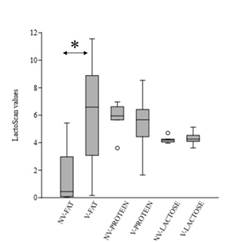
Protein content in the unvaccinated group of cows ranged from 3.61 to 6.97, compared to 1.65-8.64 in vaccinated ones. In contrast, lactose composition within vaccinated and unvaccinated groups was similar (Supplementary Fig 2). In both cases, non-significant differences were observed within cow groups (p-values >0.1). On the other hand, the fat between the two groups was statistically different (p-value = 0.003), being higher in the vaccinated group of cows (unvaccinated range:0.04-5.43; vaccinated range: 0.16-11.56) (Supplementary Fig 2).
ELISA analysis determined that colostrums presented specific anti-ClpG antibodies, although with different response levels. After establishing the positive cut-off with an Abs490 ≥0.45 (corresponding to values ≥63 % of the positivity of the technique), it was determined that 56.5 % of the colostrums (13/23) from vaccinated cows were positive for the presence of anti-ClpG antibodies. Only 14.3 % (1/7) of non-vaccinated cows were considered positive (Fig 2).
Mixed models, including variance and a median test, were used to compare the level of specific IgG against ClpG protein. It was determined that there was no variance homogeneity between both conditions (1e-04), but significant differences in medians of vaccinated and unvaccinated cows were observed (0.5735) (Fig 2).
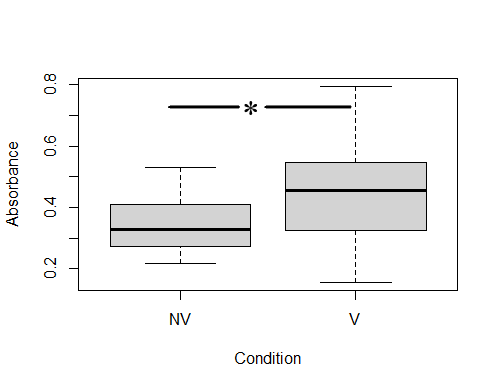
Comparison of colostrum IgG antibodies against ClpG between vaccinated and non-vaccinated cows. The number of vaccinated cows was 23, and the number of non-vaccinated cows was 9. Each colostrum was studied in triplicate. NV: non-vaccinated, V: vaccinated.
Discussion
Maternal Igs are transferred to the calves through colostrum consumption right after birth and absorbed in the small intestine of the neonate. Colostral components vary among breed, season, diet, parity, the dry period and environmental factors around the cow. Its quality is assessed by IgG content, with good quality colostrum, when it contains >50 g/L of this immunoglobulin (Altvater-Hughes et al., 2022). It is established that absorption of good quality IgG in the first 12 hours after birth is essential to achieve optimal weight gain and good health during the suckling stage, and is critical to decreasing the morbidity and mortality rates of the calves (Heinrichs et al., 2020). The mortality rate of perinatal calves in Uruguayan dairy farms due to infectious diseases is among the highest in America (Schild, 2017). This scenario requires quick and accurate actions to improve the outcome of the animals, either ensuring a better passive transfer of antibodies through a correct consumption of colostrum or a better quality and composition of the antibodies within it.
Several vaccines, with different compositions and vaccination schedules are available to prevent NCD. Different outcomes have been observed in field trials regarding NCD vaccines containing E. coli antigens, with a reduction in the incidence of diarrhea in some cases (Maier et al., 2022). Vaccines containing F5 antigen for instance have been widely applied worldwide, and although the pathogen continues to be associated with the disease, a decrease in its incidence has been observed over the years, a situation that has been attributed to the extensive use of it (Kolenda et al., 2015). Vaccination of pregnant cows between the seventh and eighth month of gestation is frequently applied in Uruguay to prevent and reduce the effect of the disease (Schild, 2017). The incidence of f5 is low in pathogenic E. coli from feces of dairy calves in Uruguay, whereas the prevalence of clpG adhesin gene is one of the highest (Caffarena et al., 2021; Umpiérrez et al., 2016, 2021). This work aimed to evaluate if vaccination of pregnant cows with a vaccine including E. coli CS31A+ bacterin induces differences in colostrum’s IgG antibodies against ClpG antigen. The colostrum composition was analyzed as a screening to determine the functional state of the mammary gland of vaccinated and unvaccinated cows. Non-significant differences were determined in the colostrum-protein or -lactose composition within vaccinated and unvaccinated cows (p-values >0.1), whereas the values were different in terms of fat (p-value = 0.003). Generally, colostrum’s fat content is higher than in milk and a large range of average fat-content, as observed in this work, has been reported (McGrath et al., 2016). Hence, it was assumed that the fat differences had no-effect on interpreting the ELISA.
The presence of anti-ClpG antibodies was detected by ELISA in both groups of bovines, which would reflect the fact that cows are naturally challenged with E. coli CS31A+ strains on dairy farms. Taking into account the values considered positive (Abs490 ≥ 0.45), the values of IgG anti-ClpG were higher in the vaccinated group of cows (56.5 % of the colostrums) compared to the unvaccinated ones (14.3 % of the colostrums), a response that would be attributed to the administration of the vaccine. Additionally, the mixed model analysis showed not variance homogeneity between groups, but significant differences in medians of the IgG values of the 23 vaccinated and the 7 unvaccinated cows. Previous works have reported the high circulation of pathogenic E. coli-CS31A+ strains in dairy herds from Uruguay (Umpiérrez et al., 2016, 2021). Given the anti-ClpG response observed in this study and the fact that it is necessary to include antigens of local circulating pathogens, the incorporation of this antigen within vaccines would improve the colostral antibody content and be helpful to reduce NCD incidence in Uruguay. Despite this, the variance differences between both groups of cows would also be attributed to intrinsic differences of the animals and the management practices. The access to dairy farms that do not vaccinate pregnant cows against NCD in Uruguay is tough, but an increase of cows in the unvaccinated group, or a controlled trial with vaccinated and unvaccianted cows from the same rodeo, should be desirable to confirm the tendency observed.
Conclusion
Vaccination of pregnant cows against NCD is frequent among dairy herds in Uruguay, and vaccines containing E. coli CS31A+ bacterin are available. This work studied the presence of IgG antibodies against the structural protein of CS31A adhesin, ClpG, in cows’ colostrum for the first time. The exposure of cows to E. coli CS31A+ strains in the herds was confirmed by the presence of specific IgG antibodies against ClpG in the colostrum of both vaccinated and unvaccinated dairy cows. The vaccination strategy showed a tendency to increase the amount of IgG antibodies against ClpG in the cows’ colostrum, which can result in a better outcome for the calves if the passive transfer of IgG effectively occurs.